Spatio-temporal tumor heterogeneity in metastatic CRC tumors: a mutational-based approach
- PMID: 30344942
- PMCID: PMC6188146
- DOI: 10.18632/oncotarget.26081
Spatio-temporal tumor heterogeneity in metastatic CRC tumors: a mutational-based approach
Abstract
It is well known that activating mutations in the KRAS and NRAS genes are associated with poor response to anti-EGFR therapies in patients with metastatic colorectal cancer (mCRC). Approximately half of the patients with wild-type (WT) KRAS colorectal carcinoma do not respond to these therapies. This could be because the treatment decision is determined by the mutational profile of the primary tumor, regardless of the presence of small tumor subclones harboring RAS mutations in lymph nodes or liver metastases. We analyzed the mutational profile of the KRAS, NRAS, BRAF and PI3KCA genes using low-density microarray technology in samples of 26 paired primary tumors, 16 lymph nodes and 34 liver metastases from 26 untreated mCRC patients (n=76 samples). The most frequent mutations found in primary tumors were KRAS (15%) and PI3KCA (15%), followed by NRAS (8%) and BRAF (4%). The distribution of the mutations in the 16 lymph node metastases analyzed was as follows: 4 (25%) in KRAS gene, 3 (19%) in NRAS gene and 1 mutation each in PI3KCA and BRAF genes (6%). As expected, the most prevalent mutation in liver metastasis was in the KRAS gene (35%), followed by PI3KCA (9%) and BRAF (6%). Of the 26 cases studied, 15 (58%) displayed an overall concordance in the mutation status detected in the lymph node metastases and liver metastases compared with primary tumor, suggesting no clonal evolution. In contrast, the mutation profiles differed in the primary tumor and lymph node/metastases samples of the remaining 11 patients (48%), suggesting a spatial and temporal clonal evolution. We confirm the presence of different mutational profiles among primary tumors, lymph node metastases and liver metastases. Our results suggest the need to perform mutational analysis in all available tumor samples of patients before deciding to commence anti-EGFR treatment.
Keywords: anti-EGFR; clonal evolution; colorectal cancer; mutational profile; tumor heterogeneity.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures
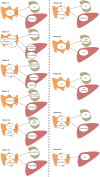
 for the primary tumor;
for the primary tumor;  lymph node;
lymph node;  liver metastasis. If more than one sample per tissue was studied it is mentioned in the figure (n= number of samples).
liver metastasis. If more than one sample per tissue was studied it is mentioned in the figure (n= number of samples).References
-
- Peeters M, Douillard JY, Van Cutsem E, Siena S, Zhang K, Williams R, Wiezorek J. Mutant KRAS codon 12 and 13 alleles in patients with metastatic colorectal cancer: assessment as prognostic and predictive biomarkers of response to panitumumab. J Clin Oncol. 2013;31:759–765. doi: 10.1200/JCO.2012.45.1492. - DOI - PubMed
-
- Rowland A, Dias MM, Wiese MD, Kichenadasse G, McKinnon RA, Karapetis CS, Sorich MJ. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Cancer. 2015;112:1888–1894. doi: 10.1038/bjc.2015.173. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

