Autophagy and unfolded protein response (UPR) regulate mammary gland involution by restraining apoptosis-driven irreversible changes
- PMID: 30345078
- PMCID: PMC6186758
- DOI: 10.1038/s41420-018-0105-y
Autophagy and unfolded protein response (UPR) regulate mammary gland involution by restraining apoptosis-driven irreversible changes
Erratum in
-
Erratum: Publisher Correction: articles initially published in wrong volume.Cell Death Discov. 2019 Jul 10;5:116. doi: 10.1038/s41420-019-0186-2. eCollection 2019. Cell Death Discov. 2019. PMID: 31312525 Free PMC article.
Abstract
The postnatal mammary gland undergoes repeated cycles of proliferation and cell death, most notably when the fully differentiated (lactating) gland dedifferentiates to a prelactation state. Accumulation of milk proteins in the secretory epithelium creates the stress signal that triggers this process (involution). How this stress is perceived, and the cellular processes that are subsequently activated, remain unclear. We now report that Unfolded Protein Response (UPR), autophagy, and apoptosis related genes cluster separately during lactation and involution in the mouse mammary gland. Time-course experiments in rodents show that autophagy and UPR signaling are tightly co-regulated at the transition from reversible to irreversible involution. Inhibition of autophagy by chloroquine or genetic deletion of one ATG7 allele enhanced progression of mammary involution into the irreversible phase, as characterized by an early/precocious induction of apoptosis. These are the first preclinical in vivo data in support of a clinical trial testing an autophagy inhibitor for prevention of intraductal breast malignancy progression to invasive breast cancer. In marked contrast, stimulation of autophagy by low dose tunicamycin treatment reduced apoptosis and extended the reversible phase of involution by sustaining the secretory epithelium. Autophagy stimulators could be used short-term to promote lactation in women experiencing difficulties or irregularities in nursing. Taken together, these data indicate that UPR and autophagy play a key role in regulating the balance between cell survival and apoptosis during normal mammary gland regression.
Conflict of interest statement
Compliance with ethical standardsThe authors declare that they have no conflict of interest.
Figures

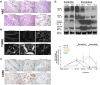


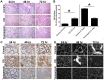
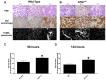
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

