Multi-Omics Profiling of the Tumor Microenvironment: Paving the Way to Precision Immuno-Oncology
- PMID: 30345255
- PMCID: PMC6182075
- DOI: 10.3389/fonc.2018.00430
Multi-Omics Profiling of the Tumor Microenvironment: Paving the Way to Precision Immuno-Oncology
Abstract
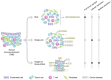
The tumor microenvironment (TME) is a multifaceted ecosystem characterized by profound cellular heterogeneity, dynamicity, and complex intercellular cross-talk. The striking responses obtained with immune checkpoint blockers, i.e., antibodies targeting immune-cell regulators to boost antitumor immunity, have demonstrated the enormous potential of anticancer treatments that target TME components other than tumor cells. However, as checkpoint blockade is currently beneficial only to a limited fraction of patients, there is an urgent need to understand the mechanisms orchestrating the immune response in the TME to guide the rational design of more effective anticancer therapies. In this Mini Review, we give an overview of the methodologies that allow studying the heterogeneity of the TME from multi-omics data generated from bulk samples, single cells, or images of tumor-tissue slides. These include approaches for the characterization of the different cell phenotypes and for the reconstruction of their spatial organization and inter-cellular cross-talk. We discuss how this broader vision of the cellular heterogeneity and plasticity of tumors, which is emerging thanks to these methodologies, offers the opportunity to rationally design precision immuno-oncology treatments. These developments are fundamental to overcome the current limitations of targeted agents and checkpoint blockers and to bring long-term clinical benefits to a larger fraction of cancer patients.
Keywords: bioinformatics; immuno-oncology; multi-omics profiling; next-generation sequencing; systems biology; systems immunology; tumor microenvironment; tumor-infiltrating immune cells.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

