Population genomic analyses of the chocolate tree, Theobroma cacao L., provide insights into its domestication process
- PMID: 30345393
- PMCID: PMC6191438
- DOI: 10.1038/s42003-018-0168-6
Population genomic analyses of the chocolate tree, Theobroma cacao L., provide insights into its domestication process
Abstract
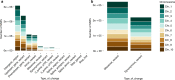
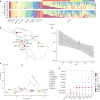
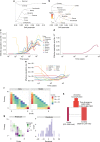
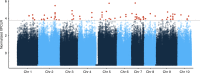
Domestication has had a strong impact on the development of modern societies. We sequenced 200 genomes of the chocolate plant Theobroma cacao L. to show for the first time to our knowledge that a single population, the Criollo population, underwent strong domestication ~3600 years ago (95% CI: 2481-13,806 years ago). We also show that during the process of domestication, there was strong selection for genes involved in the metabolism of the colored protectants anthocyanins and the stimulant theobromine, as well as disease resistance genes. Our analyses show that domesticated populations of T. cacao (Criollo) maintain a higher proportion of high-frequency deleterious mutations. We also show for the first time the negative consequences of the increased accumulation of deleterious mutations during domestication on the fitness of individuals (significant reduction in kilograms of beans per hectare per year as Criollo ancestry increases, as estimated from a GLM, P = 0.000425).
Conflict of interest statement
Co-authors Donald Livingstone III, Conrad Stack, Alberto Romero, Stefan Royaert, Osman Gutierrez, and Juan C. Motamayor are or have been employees of the funding agency MARS Inc. The authors declare no other competing financial or non-financial interests.
Figures

References
-
- Childe, G. V. Social evolution. (Watts & Co, London, 1951).
-
- Coe, S. D., Coe, M. D. & Huxtable, R. J. The true history of chocolate. (Thames and Hudson, New York, 1996).
LinkOut - more resources
Full Text Sources

