SeeVis-3D space-time cube rendering for visualization of microfluidics image data
- PMID: 30346487
- PMCID: PMC6513157
- DOI: 10.1093/bioinformatics/bty889
SeeVis-3D space-time cube rendering for visualization of microfluidics image data
Abstract
Motivation: Live cell imaging plays a pivotal role in understanding cell growth. Yet, there is a lack of visualization alternatives for quick qualitative characterization of colonies.
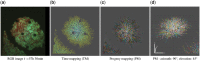
Results: SeeVis is a Python workflow for automated and qualitative visualization of time-lapse microscopy data. It automatically pre-processes the movie frames, finds particles, traces their trajectories and visualizes them in a space-time cube offering three different color mappings to highlight different features. It supports the user in developing a mental model for the data. SeeVis completes these steps in 1.15 s/frame and creates a visualization with a selected color mapping.
Availability and implementation: https://github.com/ghattab/seevis/.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2018. Published by Oxford University Press.
Figures
Similar articles
-
An automated workflow for parallel processing of large multiview SPIM recordings.Bioinformatics. 2016 Apr 1;32(7):1112-4. doi: 10.1093/bioinformatics/btv706. Epub 2015 Dec 1. Bioinformatics. 2016. PMID: 26628585 Free PMC article.
-
CellTracker: an automated toolbox for single-cell segmentation and tracking of time-lapse microscopy images.Bioinformatics. 2021 Apr 19;37(2):285-287. doi: 10.1093/bioinformatics/btaa1106. Bioinformatics. 2021. PMID: 33416830
-
dNEMO: a tool for quantification of mRNA and punctate structures in time-lapse images of single cells.Bioinformatics. 2021 May 5;37(5):677-683. doi: 10.1093/bioinformatics/btaa874. Bioinformatics. 2021. PMID: 33051642 Free PMC article.
-
SPIM workflow manager for HPC.Bioinformatics. 2019 Oct 1;35(19):3875-3876. doi: 10.1093/bioinformatics/btz140. Bioinformatics. 2019. PMID: 30799494 Free PMC article.
-
clustermq enables efficient parallelization of genomic analyses.Bioinformatics. 2019 Nov 1;35(21):4493-4495. doi: 10.1093/bioinformatics/btz284. Bioinformatics. 2019. PMID: 31134271 Free PMC article.
Cited by
-
Immersive and interactive visualization of 3D spatio-temporal data using a space time hypercube: Application to cell division and morphogenesis analysis.Front Bioinform. 2023 Mar 8;3:998991. doi: 10.3389/fbinf.2023.998991. eCollection 2023. Front Bioinform. 2023. PMID: 36969798 Free PMC article.
References
-
- Crocker J., Grier D. (1996) Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci., 179, 298–310.
-
- Klein J., et al. (2012) TLM-Tracker: software for cell segmentation, tracking and lineage analysis in time-lapse microscopy movies. Bioinformatics, 28, 2276–2277. - PubMed

