Rationalizing Secondary Pharmacology Screening Using Human Genetic and Pharmacological Evidence
- PMID: 30346593
- PMCID: PMC6358245
- DOI: 10.1093/toxsci/kfy265
Rationalizing Secondary Pharmacology Screening Using Human Genetic and Pharmacological Evidence
Abstract
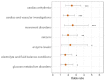
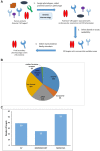
Safety-related drug failures remain a major challenge for the pharmaceutical industry. One approach to ensuring drug safety involves assessing small molecule drug specificity by examining the ability of a drug candidate to interact with a panel of "off-target" proteins, referred to as secondary pharmacology screening. Information from human genetics and pharmacology can be used to select proteins associated with adverse effects for such screening. In an analysis of marketed drugs, we found a clear relationship between the genetic and pharmacological phenotypes of a drug's off-target proteins and the observed drug side effects. In addition to using this phenotypic information for the selection of secondary pharmacology screens, we also show that it can be used to help identify drug off-target protein interactions responsible for drug-related adverse events. We anticipate that this phenotype-driven approach to secondary pharmacology screening will help to reduce safety-related drug failures due to drug off-target protein interactions.
Figures
Similar articles
-
Secondary pharmacology: screening and interpretation of off-target activities - focus on translation.Drug Discov Today. 2016 Aug;21(8):1232-42. doi: 10.1016/j.drudis.2016.04.021. Epub 2016 Apr 29. Drug Discov Today. 2016. PMID: 27140035 Review.
-
The use of biomarkers in human pharmacology (Phase I) studies.Annu Rev Pharmacol Toxicol. 2015;55:55-74. doi: 10.1146/annurev-pharmtox-011613-135918. Epub 2014 Oct 6. Annu Rev Pharmacol Toxicol. 2015. PMID: 25292425 Review.
-
The state of the art in secondary pharmacology and its impact on the safety of new medicines.Nat Rev Drug Discov. 2024 Jul;23(7):525-545. doi: 10.1038/s41573-024-00942-3. Epub 2024 May 21. Nat Rev Drug Discov. 2024. PMID: 38773351 Review.
-
Methodological innovations expand the safety pharmacology horizon.J Pharmacol Toxicol Methods. 2012 Sep;66(2):59-62. doi: 10.1016/j.vascn.2012.05.004. Epub 2012 May 20. J Pharmacol Toxicol Methods. 2012. PMID: 22617368
-
Safety pharmacology methods and regulatory considerations evolve together.J Pharmacol Toxicol Methods. 2018 Sep-Oct;93:1-6. doi: 10.1016/j.vascn.2018.06.004. Epub 2018 Jun 21. J Pharmacol Toxicol Methods. 2018. PMID: 29936032 No abstract available.
Cited by
-
Current status and future directions for a neurotoxicity hazard assessment framework that integrates in silico approaches.Comput Toxicol. 2022 May;22:100223. doi: 10.1016/j.comtox.2022.100223. Epub 2022 Mar 17. Comput Toxicol. 2022. PMID: 35844258 Free PMC article.
-
Human genetic evidence enriched for side effects of approved drugs.PLoS Genet. 2025 Mar 31;21(3):e1011638. doi: 10.1371/journal.pgen.1011638. eCollection 2025 Mar. PLoS Genet. 2025. PMID: 40163513 Free PMC article.
-
Exploration of the DARTable Genome- a Resource Enabling Data-Driven NAMs for Developmental and Reproductive Toxicity Prediction.Front Toxicol. 2022 Jan 19;3:806311. doi: 10.3389/ftox.2021.806311. eCollection 2021. Front Toxicol. 2022. PMID: 35295108 Free PMC article.
-
In silico approaches in organ toxicity hazard assessment: current status and future needs in predicting liver toxicity.Comput Toxicol. 2021 Nov;20:100187. doi: 10.1016/j.comtox.2021.100187. Epub 2021 Sep 9. Comput Toxicol. 2021. PMID: 35340402 Free PMC article.
-
A proteomic platform to identify off-target proteins associated with therapeutic modalities that induce protein degradation or gene silencing.Sci Rep. 2021 Aug 4;11(1):15856. doi: 10.1038/s41598-021-95354-3. Sci Rep. 2021. PMID: 34349202 Free PMC article.
References
-
- Abifadel M., Varret M., Rabes J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D. (2003). Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34, 154–156. - PubMed
-
- Agresti A. (2002). Categorical Data Analysis, 2nd ed.Wiley, New York, NY.
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

