Sensitive and rapid detection of TERT promoter and IDH mutations in diffuse gliomas
- PMID: 30346624
- PMCID: PMC6422442
- DOI: 10.1093/neuonc/noy167
Sensitive and rapid detection of TERT promoter and IDH mutations in diffuse gliomas
Abstract
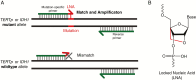
Background: Mutations in telomerase reverse transcriptase promoter (TERTp) and isocitrate dehydrogenase 1 and 2 (IDH) offer objective markers to assist in classifying diffuse gliomas into genetic subgroups. However, traditional mutation detection techniques lack sensitivity or have long turnaround times or high costs. We developed GliomaDx, an allele-specific, locked nucleic acid-based quantitative PCR assay to overcome these limitations and sensitively detect TERTp and IDH mutations.
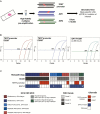
Methods: We evaluated the performance of GliomaDx on cell line DNA and frozen tissue diffuse glioma samples with variable tumor percentage to mimic use in clinical settings and validated low percentage variants using sensitive techniques including droplet digital PCR (ddPCR) and next-generation sequencing. We also developed GliomaDx Nest, which incorporates a high-fidelity multiplex pre-amplification step prior to allele-specific PCR for low-input formalin-fixed paraffin embedded (FFPE) samples.
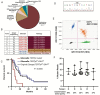
Results: GliomaDx detects the TERTp and IDH1 alterations at an analytical sensitivity of 0.1% mutant allele fraction, corresponding to 0.2% tumor cellularity. GliomaDx identified TERTp/IDH1 alterations in a cohort of frozen tissue samples with variable tumor percentage of all major diffuse glioma histologic types. GliomaDx Nest is able to detect these hotspot mutations with similar sensitivity from pre-amplified samples and was successfully tested on a cohort of clinical FFPE samples. Testing of a cohort of previously identified TERTpWT-IDHWT gliomas (by Sanger sequencing) revealed that 26.3% harbored low-percentage mutations. Analysis by ddPCR and whole exome sequencing of these tumors confirmed the low mutant fraction of these alterations and overall mutation-based tumor purity.
Conclusions: Our results show that GliomaDx can rapidly detect TERTp/IDH mutations with high sensitivity, identifying cases that might be missed due to the lack of sensitivity of other techniques. This approach may facilitate more objective classification of diffuse glioma samples in clinical settings such as intraoperative diagnosis or in testing cases with low tumor purity.
Keywords: IDH mutation; TERT promoter mutation; glioma classification; qPCR; sequencing.
© The Author(s) 2018. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


Comment in
-
TERT promoter mutation as a diagnostic marker for diffuse gliomas.Neuro Oncol. 2019 Mar 18;21(4):417-418. doi: 10.1093/neuonc/noz025. Neuro Oncol. 2019. PMID: 30852620 Free PMC article. No abstract available.
References
-
- McLendon RE. Errors in surgical neuropathology and the influence of cognitive biases: the psychology of intelligence analysis. Arch Pathol Lab Med. 2006;130(5):613–616. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

