Synthesis of Tripeptide Derivatives with Three Stereogenic Centers and Chiral Recognition Probed by Tetraaza Macrocyclic Chiral Solvating Agents Derived from d-Phenylalanine and (1 S,2 S)-(+)-1,2-Diaminocyclohexane via 1H NMR Spectroscopy
- PMID: 30346768
- PMCID: PMC6499380
- DOI: 10.1021/acs.joc.8b02212
Synthesis of Tripeptide Derivatives with Three Stereogenic Centers and Chiral Recognition Probed by Tetraaza Macrocyclic Chiral Solvating Agents Derived from d-Phenylalanine and (1 S,2 S)-(+)-1,2-Diaminocyclohexane via 1H NMR Spectroscopy
Abstract
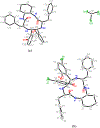
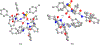
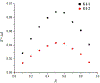
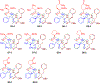
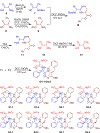
Enantiomers of a series of tripeptide derivatives with three stereogenic centers (±)-G1-G9 have been prepared from d- and l-α-amino acids as guests for chiral recognition by 1H NMR spectroscopy. In the meantime, a family of tetraaza macrocyclic chiral solvating agents (TAMCSAs) 1a-1d has been synthesized from d-phenylalanine and (1 S,2 S)-(+)-1,2-diaminocyclohexane. Discrimination of enantiomers of (±)-G1-G9 was carried out in the presence of TAMCSAs 1a-1d by 1H NMR spectroscopy. The results indicate that enantiomers of (±)-G1-G9 can be effectively discriminated in the presence of TAMCSAs 1a-1d by 1H NMR signals of multiple protons exhibiting nonequivalent chemical shifts (ΔΔδ) up to 0.616 ppm. Furthermore, enantiomers of (±)-G1-G9 were easily assigned by comparing 1H NMR signals of the split corresponding protons with those attributed to a single enantiomer. Different optical purities (ee up to 90%) of G1 were clearly observed and calculated in the presence of TAMCSAs 1a-1d, respectively. Intermolecular hydrogen bonding interactions were demonstrated through theoretical calculations of enantiomers of (±)-G1 with TAMCSA 1a by means of the hybrid functional theory with the standard basis sets of 3-21G of the Gaussian 03 program.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
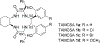



Similar articles
-
Chiral Recognition of Chiral (Hetero)Cyclic Derivatives Probed by Tetraaza Macrocyclic Chiral Solvating Agents via 1H NMR Spectroscopy.Anal Chem. 2024 Apr 2;96(13):5188-5194. doi: 10.1021/acs.analchem.3c05395. Epub 2024 Mar 20. Anal Chem. 2024. PMID: 38506628 Free PMC article.
-
Chiral Recognition of Hydantoin Derivatives Enabled by Tetraaza Macrocyclic Chiral Solvating Agents Using 1H NMR Spectroscopy.J Org Chem. 2022 Jun 17;87(12):7934-7944. doi: 10.1021/acs.joc.2c00587. Epub 2022 Jun 8. J Org Chem. 2022. PMID: 35675642 Free PMC article.
-
Chiral discrimination of α-hydroxy acids and N-Ts-α-amino acids induced by tetraaza macrocyclic chiral solvating agents by using 1H NMR spectroscopy.Org Biomol Chem. 2017 Feb 21;15(7):1642-1650. doi: 10.1039/c6ob02578a. Epub 2017 Jan 27. Org Biomol Chem. 2017. PMID: 28127599 Free PMC article.
-
Discrimination of Enantiomers of Dipeptide Derivatives with Two Chiral Centers by Tetraaza Macrocyclic Chiral Solvating Agents Using 1H NMR Spectroscopy.Org Chem Front. 2016 Dec 1;3(12):1716-1724. doi: 10.1039/C6QO00521G. Epub 2016 Sep 30. Org Chem Front. 2016. PMID: 28191319 Free PMC article.
-
Enantiotopic discrimination in the NMR spectrum of prochiral solutes in chiral liquid crystals.Chem Soc Rev. 2015 Apr 21;44(8):2330-75. doi: 10.1039/c4cs00260a. Chem Soc Rev. 2015. PMID: 25676790 Review.
Cited by
-
Chiral Recognition of Chiral (Hetero)Cyclic Derivatives Probed by Tetraaza Macrocyclic Chiral Solvating Agents via 1H NMR Spectroscopy.Anal Chem. 2024 Apr 2;96(13):5188-5194. doi: 10.1021/acs.analchem.3c05395. Epub 2024 Mar 20. Anal Chem. 2024. PMID: 38506628 Free PMC article.
-
Efficient Enantiodifferentiation of Carboxylic Acids Using BINOL-Based Amino Alcohol as a Chiral NMR Solvating Agent.Front Chem. 2020 May 4;8:336. doi: 10.3389/fchem.2020.00336. eCollection 2020. Front Chem. 2020. PMID: 32432082 Free PMC article.
-
Chiral Recognition of Hydantoin Derivatives Enabled by Tetraaza Macrocyclic Chiral Solvating Agents Using 1H NMR Spectroscopy.J Org Chem. 2022 Jun 17;87(12):7934-7944. doi: 10.1021/acs.joc.2c00587. Epub 2022 Jun 8. J Org Chem. 2022. PMID: 35675642 Free PMC article.
-
Azaheterocyclic diphenylmethanol chiral solvating agents for the NMR chiral discrimination of alpha-substituted carboxylic acids.RSC Adv. 2020 Sep 18;10(57):34605-34611. doi: 10.1039/d0ra06312f. eCollection 2020 Sep 16. RSC Adv. 2020. PMID: 35514411 Free PMC article.
References
-
- Pirkle WH; Pochapsky TC Consideration of Chiral Recognition Relevant to the Liquid Chromatographic Separation of Enantiomers. Chem. Rev 1989, 89, 347–362.
- Tachibana K; Ohnishi A Reversed-Phase Liquid Chromatographic Separation of Enantiomers on Polysaccharide Type Chiral Stationary Phase. J. Chromatogr. A 2001, 906, 127–154. - PubMed
- Ema T; Hamada K; Sugita K; Nagata Y; Sakai T; Ohnishi A Synthesis and Evaluation of Chiral Selectors with Multiple Hydrogen-Bonding Sites in the Macrocyclic Cavities. J. Org. Chem 2010, 75, 4492–4500. - PubMed
- Borowiecki P Enantiodifferentiation of Promethazine Using (S)-(–)-BINOL as the NMR Chiral Solvating Agent: Determination of the Enantiomeric Purity and Performance Comparison with Traditional Chiral HPLC. Tetrahedron: Asymmetry 2015, 26, 16–23.
-
- Nieto S; Lynch VM; Anslyn EV; Kim H; Chin J Rapid Enantiomeric Excess and Concentration Determination Using Simple Racemic Metal Complexes. Org. Lett 2008, 10, 5167–5170. - PMC - PubMed
- Joyce LA; Maynor MS; Dragna JM; da Cruz GM; Lynch VM; Canary JW; Anslyn EV A Simple Method for the Determination of Enantiomeric Excess and Identity of Chiral Carboxylic Acids. J. Am. Chem. Soc 2011, 133, 13746–13752. - PMC - PubMed
- Zardi P; Wurst K; Licini G; Zonta C Concentration-Independent Stereodynamic g-Probe for Chiroptical Enantiomeric Excess Determination. J. Am. Chem. Soc 2017, 139, 15616–15619. - PubMed
-
- Czerwenka C; Lindner W Enantiomer Discrimination of Peptides by Tandem Mass Spectroscopy: Influence of the Peptide Sequence on Chiral Recognition. Rapid Commun. Mass Spectrom 2004, 18, 2713–2718. - PubMed
- Ravikumar M; Prabhakar S; Vairamani M Chiral Discrimination of α-Amino Acids by the DNA Triplet GCA. Chem. Commun 2007, 392–394. - PubMed
- Faggi E; Vicent C; Luis SV; Alfonso I Stereoselective Recognition of the Ac-Glu-Tyr-OH Dipeptide by Pseudopeptidic Cages. Org. Biomol. Chem 2015, 13, 11721–11731. - PubMed
-
- Parker D NMR Determination of Enantiomeric Purity. Chem. Rev 1991, 91, 1441–1457.
- Wenzel TJ; Wilcox JD Chiral Reagents for the Determination of Enantiomeric Excess and Absolute Configuration Using NMR Spectroscopy. Chirality 2003, 15, 256–270. - PubMed
- Seco JM; Quiñoa E; Riguera R The Assignment of́ Abosolute Configuration by NMR. Chem. Rev 2004, 104, 17–117. - PubMed
- Ye XX; Lei XX; Chen ZF; Zhang LX; Zhang AJ In Situ Measurement of the Enantiomeric Excess of Alcohols and Amines under Asymmetric Reduction Reaction by 1H NMR. Org. Lett 2010, 12, 3238–3241. - PubMed
- Labuta J; Ishihara S; Šikorsky T; Futera Z;́ Shundo A; Hanykova L; Burda JV; Ariga K; Hill JP NMŔ Spectroscopic Detection of Chirality and Enantiopurity in Referenced Systems without Formation of Diastereomers. Nat. Commun 2013, 4, 2188. - PMC - PubMed
- Labuta J; Hill JP; Ishihara S; Hanykova L; Ariga K Chiral Sensing by Nonchiral Tetrapyrroles. Acc. Chem. Res 2015, 48, 521–529. - PubMed
- Bian GL; Yang SW; Huang HY; Zong H; Song L; Fan HJ; Sun XQ Chirality Sensing of Tertiary Alcohols by a Novel Strong Hydrogen-Bonding Donor–Selenourea. Chem. Sci 2016, 7, 932–938. - PMC - PubMed
- Ito S; Okuno M; Asami M Differentiation of Enantiomeric Anions by NMR Spectroscopy with Chiral Bisurea Receptors. Org. Biomol. Chem 2018, 16, 213–222. - PubMed
- Ishihara S; Labuta J; Futera Z; Mori S; Sato H; Ariga K; Hill JP NMR Spectroscopic Determination of Enantiomeric Excess Using Small Prochiral Molecules. J. Phys. Chem. B 2018, 122, 5114–5120. - PubMed
-
- Tsubaki K; Tanima D; Nuruzzaman M; Kusumoto T; Fuji K; Kawabata T Visual Enantiomeric Recognition of Amino Acid Derivatives in Protic Solvents. J. Org. Chem 2005, 70, 4609–4616. - PubMed
- Lee CS; Teng PF; Wong WL; Kwong HL; Chan ASC New C2-Symmetric 2,2’-Bipyridine Crown Macrocycles for Enantioselective Recognition of Amino Acid Derivatives. Tetrahedron 2005, 61, 7924–7930.
- Demirtas HN; Bozkurt S; Durmaz M; Yilmaz M; Sirit A Chiral Calix[4]azacrowns for Enantiomeric Recognition of Amino Acid Derivatives. Tetrahedron 2009, 65, 3014–3018.
- Cheng C; Cai ZW; Peng XS; Wong HNC Enantiomeric Recognition of Amino Acid Salts by Macrocyclic Crown Ethers Derived from Enantiomerically Pure 1,8,9,16-Tetrahydroxytetraphenylenes. J. Org. Chem 2013, 78, 8562–8573. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

