Bone marrow derived-mesenchymal stem cells downregulate IL17A dependent IL6/STAT3 signaling pathway in CCl4-induced rat liver fibrosis
- PMID: 30346985
- PMCID: PMC6197688
- DOI: 10.1371/journal.pone.0206130
Bone marrow derived-mesenchymal stem cells downregulate IL17A dependent IL6/STAT3 signaling pathway in CCl4-induced rat liver fibrosis
Abstract
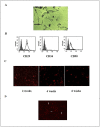
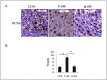
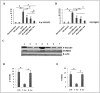
Therapeutic potential of bone marrow-derived mesenchymal stem cells (BM-MSCs) has been reported in several animal models of liver fibrosis. Interleukin (IL) 17A, IL6 and Stat3 have been described to play crucial roles in chronic liver injury. However, the modulatory effect of MSCs on these markers was controversial in different diseases. BM-MSCs might activate the IL6/STAT3 signaling pathway and promote cell invasion in hepatocellular carcinoma, but the immunomodulatory role of BM-MSCs on IL17A/IL6/STAT3 was not fully elucidated in liver fibrosis. In the present study, we evaluated the capacity of the BM-MSCs in the modulation of cytokines milieu and signal transducers, based on unique inflammatory genes Il17a and Il17f and their receptors Il17rc and their effect on the IL6/STAT3 pathway in CCl4-induced liver fibrosis in rats. A single dose of BM-MSCs was administered to the group with induced liver fibrosis, and the genes and proteins of interest were evaluated along six weeks after treatment. Our results showed a significant downregulation of Il17a, Il17ra, il17f and Il17rc genes. In accordance, BM-MSCs administration declined IL17, IL2 and IL6 serum proteins and downregulated IL17A and IL17RA proteins in liver tissue. Interestingly, BM-MSCs downregulated both Stat3 mRNA expression and p-STAT3, while Stat5a gene was downregulated and p-STAT5 protein was elevated. Also P-SMAD3 and TGFβR2 proteins were downregulated in response to BM-MSCs treatment. Collectively, we suggest that BM-MSCs might play an immunomodulatory role in the treatment of liver fibrosis through downregulation of IL17A affecting IL6/STAT3 signaling pathway.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- Friedman S. Liver fibrosis: from bench to bedside. Journal of hepatology Supplement. 2003;38(1):38–53. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

