Transcription apparatus of the yeast virus-like elements: Architecture, function, and evolutionary origin
- PMID: 30346988
- PMCID: PMC6211774
- DOI: 10.1371/journal.ppat.1007377
Transcription apparatus of the yeast virus-like elements: Architecture, function, and evolutionary origin
Abstract
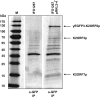
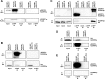
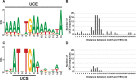
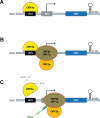
Extrachromosomal hereditary elements such as organelles, viruses, and plasmids are important for the cell fitness and survival. Their transcription is dependent on host cellular RNA polymerase (RNAP) or intrinsic RNAP encoded by these elements. The yeast Kluyveromyces lactis contains linear cytoplasmic DNA virus-like elements (VLEs, also known as linear plasmids) that bear genes encoding putative non-canonical two-subunit RNAP. Here, we describe the architecture and identify the evolutionary origin of this transcription machinery. We show that the two RNAP subunits interact in vivo, and this complex interacts with another two VLE-encoded proteins, namely the mRNA capping enzyme and a putative helicase. RNAP, mRNA capping enzyme and the helicase also interact with VLE-specific DNA in vivo. Further, we identify a promoter sequence element that causes 5' mRNA polyadenylation of VLE-specific transcripts via RNAP slippage at the transcription initiation site, and structural elements that precede the termination sites. As a result, we present a first model of the yeast virus-like element transcription initiation and intrinsic termination. Finally, we demonstrate that VLE RNAP and its promoters display high similarity to poxviral RNAP and promoters of early poxviral genes, respectively, thereby pointing to their evolutionary origin.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Autoselection of cytoplasmic yeast virus like elements encoding toxin/antitoxin systems involves a nuclear barrier for immunity gene expression.PLoS Genet. 2015 May 14;11(5):e1005005. doi: 10.1371/journal.pgen.1005005. eCollection 2015 May. PLoS Genet. 2015. PMID: 25973601 Free PMC article.
-
In vitro promoter recognition by the catalytic subunit of plant phage-type RNA polymerases.Plant Mol Biol. 2016 Oct;92(3):357-69. doi: 10.1007/s11103-016-0518-z. Epub 2016 Aug 6. Plant Mol Biol. 2016. PMID: 27497992 Free PMC article.
-
A novel bacteriophage-encoded RNA polymerase binding protein inhibits transcription initiation and abolishes transcription termination by host RNA polymerase.J Mol Biol. 2002 Jun 28;320(1):11-22. doi: 10.1016/S0022-2836(02)00420-5. J Mol Biol. 2002. PMID: 12079331
-
Structure and function of archaeal RNA polymerases.Mol Microbiol. 2007 Sep;65(6):1395-404. doi: 10.1111/j.1365-2958.2007.05876.x. Epub 2007 Aug 14. Mol Microbiol. 2007. PMID: 17697097 Review.
-
The evolutionary conservation of eukaryotic gene transcription.Experientia. 1989 Oct 15;45(10):972-83. doi: 10.1007/BF01953055. Experientia. 1989. PMID: 2680577 Review.
Cited by
-
The African Swine Fever Virus Transcriptome.J Virol. 2020 Apr 16;94(9):e00119-20. doi: 10.1128/JVI.00119-20. Print 2020 Apr 16. J Virol. 2020. PMID: 32075923 Free PMC article.
-
The role of plasmid copy number and mutation rate in evolutionary outcomes.Nat Ecol Evol. 2025 Jul 14. doi: 10.1038/s41559-025-02792-7. Online ahead of print. Nat Ecol Evol. 2025. PMID: 40659873
-
Transcriptome view of a killer: African swine fever virus.Biochem Soc Trans. 2020 Aug 28;48(4):1569-1581. doi: 10.1042/BST20191108. Biochem Soc Trans. 2020. PMID: 32725217 Free PMC article. Review.
-
The replication machinery of LUCA: common origin of DNA replication and transcription.BMC Biol. 2020 Jun 9;18(1):61. doi: 10.1186/s12915-020-00800-9. BMC Biol. 2020. PMID: 32517760 Free PMC article.
-
Structure of the recombinant RNA polymerase from African Swine Fever Virus.Nat Commun. 2024 Feb 21;15(1):1606. doi: 10.1038/s41467-024-45842-7. Nat Commun. 2024. PMID: 38383525 Free PMC article.
References
-
- Jeske S, Meinhardt F, Klassen R. Extranuclear Inheritance: Virus-Like DNA-Elements in Yeast In: Esser K, Löttge U, Beyschlag W, Murata J, editors. Progress in Botany. 68: Springer, Berlin, Heidelberg; 2007. p. 98–129.
-
- Stam JC, Kwakman J, Meijer M, Stuitje AR. Efficient isolation of the linear DNA killer plasmid of Kluyveromyces lactis: evidence for location and expression in the cytoplasm and characterization of their terminally bound proteins. Nucleic Acids Res. 1986;14(17):6871–6884. 10.1093/nar/14.17.6871 . - DOI - PMC - PubMed
-
- Sor F, Fukuhara H. Structure of a linear plasmid of the yeast Kluyveromyces lactis; Compact organization of the killer genome. Curr Genet. 1985;9(2):147–155. 10.1007/Bf00436963 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

