Photochemical Rescue of a Conformationally Inactivated Ribonucleotide Reductase
- PMID: 30347141
- PMCID: PMC6249109
- DOI: 10.1021/jacs.8b07902
Photochemical Rescue of a Conformationally Inactivated Ribonucleotide Reductase
Abstract
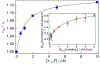
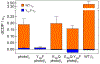
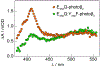
Class Ia ribonucleotide reductase (RNR) of Escherichia coli contains an unusually stable tyrosyl radical cofactor in the β2 subunit (Y122•) necessary for nucleotide reductase activity. Upon binding the cognate α2 subunit, loaded with nucleoside diphosphate substrate and an allosteric/activity effector, a rate determining conformational change(s) enables rapid radical transfer (RT) within the active α2β2 complex from the Y122• site in β2 to the substrate activating cysteine residue (C439) in α2 via a pathway of redox active amino acids (Y122[β] ↔ W48[β]? ↔ Y356[β] ↔ Y731[α] ↔ Y730[α] ↔ C439[α]) spanning >35 Å. Ionizable residues at the α2β2 interface are essential in mediating RT, and therefore control activity. One of these mutations, E350X (where X = A, D, Q) in β2, obviates all RT, though the mechanism of control by which E350 mediates RT remains unclear. Herein, we utilize an E350Q-photoβ2 construct to photochemically rescue RNR activity from an otherwise inactive construct, wherein the initial RT event (Y122• → Y356) is replaced by direct photochemical radical generation of Y356•. These data present compelling evidence that E350 conveys allosteric information between the α2 and β2 subunits facilitating conformational gating of RT that specifically targets Y122• reduction, while the fidelity of the remainder of the RT pathway is retained.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
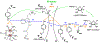
Similar articles
-
Glutamate 350 Plays an Essential Role in Conformational Gating of Long-Range Radical Transport in Escherichia coli Class Ia Ribonucleotide Reductase.Biochemistry. 2017 Feb 14;56(6):856-868. doi: 10.1021/acs.biochem.6b01145. Epub 2017 Feb 2. Biochemistry. 2017. PMID: 28103007 Free PMC article.
-
Reversible, long-range radical transfer in E. coli class Ia ribonucleotide reductase.Acc Chem Res. 2013 Nov 19;46(11):2524-35. doi: 10.1021/ar4000407. Epub 2013 Jun 4. Acc Chem Res. 2013. PMID: 23730940 Free PMC article.
-
A >200 meV Uphill Thermodynamic Landscape for Radical Transport in Escherichia coli Ribonucleotide Reductase Determined Using Fluorotyrosine-Substituted Enzymes.J Am Chem Soc. 2016 Oct 19;138(41):13706-13716. doi: 10.1021/jacs.6b08200. Epub 2016 Oct 7. J Am Chem Soc. 2016. PMID: 28068088 Free PMC article.
-
The prototypic class Ia ribonucleotide reductase from Escherichia coli: still surprising after all these years.Biochem Soc Trans. 2012 Jun 1;40(3):523-30. doi: 10.1042/BST20120081. Biochem Soc Trans. 2012. PMID: 22616862 Free PMC article. Review.
-
Long-range proton-coupled electron transfer in the Escherichia coli class Ia ribonucleotide reductase.Essays Biochem. 2017 May 9;61(2):281-292. doi: 10.1042/EBC20160072. Print 2017 May 9. Essays Biochem. 2017. PMID: 28487404 Review.
Cited by
-
Radical Transport Facilitated by a Proton Transfer Network at the Subunit Interface of Ribonucleotide Reductase.J Am Chem Soc. 2023 Mar 8;145(9):5145-5154. doi: 10.1021/jacs.2c11483. Epub 2023 Feb 22. J Am Chem Soc. 2023. PMID: 36812162 Free PMC article.
-
Structure of a trapped radical transfer pathway within a ribonucleotide reductase holocomplex.Science. 2020 Apr 24;368(6489):424-427. doi: 10.1126/science.aba6794. Epub 2020 Mar 26. Science. 2020. PMID: 32217749 Free PMC article.
-
Kinetic model for reversible radical transfer in ribonucleotide reductase.Proc Natl Acad Sci U S A. 2022 Jun 21;119(25):e2202022119. doi: 10.1073/pnas.2202022119. Epub 2022 Jun 17. Proc Natl Acad Sci U S A. 2022. PMID: 35714287 Free PMC article.
-
19F Electron-Nuclear Double Resonance Reveals Interaction between Redox-Active Tyrosines across the α/β Interface of E. coli Ribonucleotide Reductase.J Am Chem Soc. 2022 Jun 29;144(25):11270-11282. doi: 10.1021/jacs.2c02906. Epub 2022 Jun 2. J Am Chem Soc. 2022. PMID: 35652913 Free PMC article.
-
Gated Proton Release during Radical Transfer at the Subunit Interface of Ribonucleotide Reductase.J Am Chem Soc. 2021 Jan 13;143(1):176-183. doi: 10.1021/jacs.0c07879. Epub 2020 Dec 23. J Am Chem Soc. 2021. PMID: 33353307 Free PMC article.
References
-
- Stubbe J and van der Donk W Ribonucleotide reductases: radical enzymes with suicidal tendencies. Chem. Biol 1995. 2, 793–801. - PubMed
-
- Jordan A and Reichard P Ribonucleotide reductases. Annu. Rev. Biochem 1998, 67, 71–98. - PubMed
-
- Stubbe J and Ackles D On the mechanism of ribonucleoside diphosphate reductase from Escherichia coli. Evidence for 3’-C-H bond cleavage. J. Biol. Chem 1980, 255, 8027–8030. - PubMed
-
- Licht S, Gerfen GJ, and Stubbe J Thiyl radicals in ribonucleotide reductase. Science 1996, 271, 477–481. - PubMed
-
- Licht S and Stubbe J Mechanistic investigations of ribonucleotide reductases. Comp. Nat. Prod. Chem 1999, 5, 163–203.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

