Sequential specification of oligodendrocyte lineage cells by distinct levels of Hedgehog and Notch signaling
- PMID: 30347186
- PMCID: PMC6263812
- DOI: 10.1016/j.ydbio.2018.10.004
Sequential specification of oligodendrocyte lineage cells by distinct levels of Hedgehog and Notch signaling
Abstract
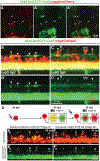
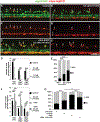
During development of the central nervous system oligodendrocyte precursor cells (OPCs) give rise to both myelinating oligodendrocytes and NG2 glia, which are the most proliferative cells in the adult mammalian brain. NG2 glia retain characteristics of OPCs, and some NG2 glia produce oligodendrocytes, but many others persist throughout adulthood. Why some OPCs differentiate as oligodendrocytes during development whereas others persist as OPCs and acquire characteristics of NG2 glia is not known. Using zebrafish spinal cord as a model, we found that OPCs that differentiate rapidly as oligodendrocytes and others that remain as OPCs arise in sequential waves from distinct neural progenitors. Additionally, oligodendrocyte and persistent OPC fates are specified during a defined critical period by small differences in Shh signaling and Notch activity, which modulates Shh signaling response. Thus, our data indicate that OPCs fated to produce oligodendrocytes or remain as OPCs during development are specified as distinct cell types, raising the possibility that the myelinating potential of OPCs is set by graded Shh signaling activity.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures






References
-
- Agius E, Soukkarieh C, Danesin C, Kan P, Takebayashi H, Soula C, and Cochard P (2004). Converse control of oligodendrocyte and astrocyte lineage development by Sonic hedgehog in the chick spinal cord. Dev. Biol 270, 308–321. - PubMed
-
- Barth KA, and Wilson SW (1995). Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development 121, 1755–1768. - PubMed
-
- Bussmann J, and Schulte-Merker S (2011a). Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development 138, 4327–4332. - PubMed
-
- Bussmann J, and Schulte-Merker S (2011b). Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development 138, 4327–4332. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

