Distinct Signatures of Host Defense Suppression by Plant-Feeding Mites
- PMID: 30347842
- PMCID: PMC6214137
- DOI: 10.3390/ijms19103265
Distinct Signatures of Host Defense Suppression by Plant-Feeding Mites
Abstract
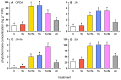
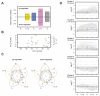
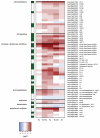
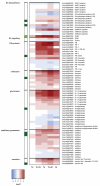
Tomato plants are attacked by diverse herbivorous arthropods, including by cell-content-feeding mites, such as the extreme generalist Tetranychus urticae and specialists like Tetranychus evansi and Aculops lycopersici. Mite feeding induces plant defense responses that reduce mite performance. However, T. evansi and A. lycopersici suppress plant defenses via poorly understood mechanisms and, consequently, maintain a high performance on tomato. On a shared host, T. urticae can be facilitated by either of the specialist mites, likely due to the suppression of plant defenses. To better understand defense suppression and indirect plant-mediated interactions between herbivorous mites, we used gene-expression microarrays to analyze the transcriptomic changes in tomato after attack by either a single mite species (T. urticae, T. evansi, A. lycopersici) or two species simultaneously (T. urticae plus T. evansi or T. urticae plus A. lycopersici). Additionally, we assessed mite-induced changes in defense-associated phytohormones using LC-MS/MS. Compared to non-infested controls, jasmonates (JAs) and salicylate (SA) accumulated to higher amounts upon all mite-infestation treatments, but the response was attenuated after single infestations with defense-suppressors. Strikingly, whereas 8 to 10% of tomato genes were differentially expressed upon single infestations with T. urticae or A. lycopersici, respectively, only 0.1% was altered in T. evansi-infested plants. Transcriptome analysis of dual-infested leaves revealed that A. lycopersici primarily suppressed T. urticae-induced JA defenses, while T. evansi dampened T. urticae-triggered host responses on a transcriptome-wide scale. The latter suggests that T. evansi not solely down-regulates plant gene expression, but rather directs it back towards housekeeping levels. Our results provide valuable new insights into the mechanisms underlying host defense suppression and the plant-mediated facilitation of competing herbivores.
Keywords: comparative transcriptomics; defense suppression; dual infestation; facilitation; herbivore; plant defense; plant-mediated interactions; tomato red spider mite (Tetranychus evansi); tomato russet mite (Aculops lycopersici); two-spotted spider mite (Tetranychus urticae).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Overcompensation of herbivore reproduction through hyper-suppression of plant defenses in response to competition.New Phytol. 2017 Jun;214(4):1688-1701. doi: 10.1111/nph.14543. Epub 2017 Apr 7. New Phytol. 2017. PMID: 28386959 Free PMC article.
-
Spatiotemporal heterogeneity of tomato induced defense responses affects spider mite performance and behavior.Plant Signal Behav. 2017 Oct 3;12(10):e1370526. doi: 10.1080/15592324.2017.1370526. Epub 2017 Aug 31. Plant Signal Behav. 2017. PMID: 28857667 Free PMC article.
-
Defense suppression benefits herbivores that have a monopoly on their feeding site but can backfire within natural communities.BMC Biol. 2014 Nov 18;12:98. doi: 10.1186/s12915-014-0098-9. BMC Biol. 2014. PMID: 25403155 Free PMC article.
-
Why Do Herbivorous Mites Suppress Plant Defenses?Front Plant Sci. 2018 Jul 30;9:1057. doi: 10.3389/fpls.2018.01057. eCollection 2018. Front Plant Sci. 2018. PMID: 30105039 Free PMC article. Review.
-
Plant-Herbivore Interactions: A Case of an Extreme Generalist, the Two-Spotted Spider Mite Tetranychus urticae.Mol Plant Microbe Interact. 2017 Dec;30(12):935-945. doi: 10.1094/MPMI-07-17-0168-CR. Epub 2017 Oct 23. Mol Plant Microbe Interact. 2017. PMID: 28857675 Review.
Cited by
-
Early Molecular Responses of Tomato to Combined Moderate Water Stress and Tomato Red Spider Mite Tetranychus evansi Attack.Plants (Basel). 2020 Aug 31;9(9):1131. doi: 10.3390/plants9091131. Plants (Basel). 2020. PMID: 32878349 Free PMC article.
-
Genome streamlining in a minute herbivore that manipulates its host plant.Elife. 2020 Oct 23;9:e56689. doi: 10.7554/eLife.56689. Elife. 2020. PMID: 33095158 Free PMC article.
-
Comparison of Tomato Transcriptomic Profiles Reveals Overlapping Patterns in Abiotic and Biotic Stress Responses.Int J Mol Sci. 2023 Feb 17;24(4):4061. doi: 10.3390/ijms24044061. Int J Mol Sci. 2023. PMID: 36835470 Free PMC article.
-
Role of LEAFLESS, an AP2/ERF family transcription factor, in the regulation of trichome specialized metabolism.New Phytol. 2025 Jul;247(2):774-790. doi: 10.1111/nph.70198. Epub 2025 May 21. New Phytol. 2025. PMID: 40400206 Free PMC article.
-
Transcriptional and metabolite analysis reveal a shift in direct and indirect defences in response to spider-mite infestation in cucumber (Cucumis sativus).Plant Mol Biol. 2020 Jul;103(4-5):489-505. doi: 10.1007/s11103-020-01005-y. Epub 2020 Apr 18. Plant Mol Biol. 2020. PMID: 32306368 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- ALW TTI Green Genetics 828.08.001/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- ALW TOP 854.11.005/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- ALW 'Meer met minder' 847.13.005/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- STW VIDI-13492/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- 12T9818N/Fonds Wetenschappelijk Onderzoek
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

