In vivo competition and horizontal gene transfer among distinct Staphylococcus aureus lineages as major drivers for adaptational changes during long-term persistence in humans
- PMID: 30348081
- PMCID: PMC6198438
- DOI: 10.1186/s12866-018-1308-3
In vivo competition and horizontal gene transfer among distinct Staphylococcus aureus lineages as major drivers for adaptational changes during long-term persistence in humans
Abstract
Background: The airways of the majority of adolescent cystic fibrosis (CF) patients are persistently colonized or infected by Staphylococcus aureus. Using whole genome sequencing, we studied the evolutionary traits within a S. aureus population in the airways of a CF patient hypothesizing that horizontal gene transfer (HGT) and inter-bacterial interaction play a major role in adaptation during long-term persistence.
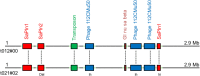
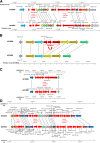
Results: Whole genome sequencing of 21 S. aureus isolates spanning 13 years resulted in seven lineages defined by the spa types t012, t021, t331, t338, t364, t056, and t2351. Of these, the successfully persisting lineages t012 and t021 were closely related suggesting the evolution of t021 from t012, which was further corroborated by a nearly identical, syntenic set of mobile genetic elements. During transformation from t012 to t021, an increase of genomic changes including HGT from other S. aureus lineages was detected.
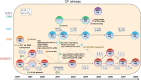
Conclusions: In summary, our in vivo data enabled us to conceptualize an evolutionary model showing the impact of HGT and inter-bacterial interaction on bacterial long-term adaptation to the human host during CF.
Keywords: Adaptation; Cystic fibrosis; Genome sequencing; Horizontal gene transfer; Staphylococcus aureus.
Conflict of interest statement
Ethics approval and consent to participate
The study was approved by the Ethical Committee of the Aerztekammer Westfalen-Lippe and of the Medical Faculty, University of Muenster (vote no. 2010–155-f-S). The need for consent was waived in this vote.
Consent for publication
There was no need for a consent for publication (see ethics approval and consent to participate).
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Transcriptional adaptations during long-term persistence of Staphylococcus aureus in the airways of a cystic fibrosis patient.Int J Med Microbiol. 2015 Jan;305(1):38-46. doi: 10.1016/j.ijmm.2014.10.005. Epub 2014 Oct 29. Int J Med Microbiol. 2015. PMID: 25439320
-
Extended Staphylococcus aureus persistence in cystic fibrosis is associated with bacterial adaptation.Int J Med Microbiol. 2013 Dec;303(8):685-92. doi: 10.1016/j.ijmm.2013.09.012. Epub 2013 Oct 8. Int J Med Microbiol. 2013. PMID: 24183484
-
Polyclonality, Shared Strains, and Convergent Evolution in Chronic Cystic Fibrosis Staphylococcus aureus Airway Infection.Am J Respir Crit Care Med. 2021 May 1;203(9):1127-1137. doi: 10.1164/rccm.202003-0735OC. Am J Respir Crit Care Med. 2021. PMID: 33296290 Free PMC article.
-
Whole genome sequencing of bacteria in cystic fibrosis as a model for bacterial genome adaptation and evolution.Expert Rev Anti Infect Ther. 2014 Mar;12(3):343-55. doi: 10.1586/14787210.2014.887441. Epub 2014 Feb 6. Expert Rev Anti Infect Ther. 2014. PMID: 24502835 Review.
-
Staphylococcus aureus genomics and the impact of horizontal gene transfer.Int J Med Microbiol. 2014 Mar;304(2):103-9. doi: 10.1016/j.ijmm.2013.11.010. Epub 2013 Dec 1. Int J Med Microbiol. 2014. PMID: 24439196 Review.
Cited by
-
Niche-specific genome degradation and convergent evolution shaping Staphylococcus aureus adaptation during severe infections.Elife. 2022 Jun 14;11:e77195. doi: 10.7554/eLife.77195. Elife. 2022. PMID: 35699423 Free PMC article.
-
Application of magnetic nanoparticles in nucleic acid detection.J Nanobiotechnology. 2020 Apr 21;18(1):62. doi: 10.1186/s12951-020-00613-6. J Nanobiotechnology. 2020. PMID: 32316985 Free PMC article. Review.
-
Staphylococcus aureus and Cystic Fibrosis-A Close Relationship. What Can We Learn from Sequencing Studies?Pathogens. 2021 Sep 13;10(9):1177. doi: 10.3390/pathogens10091177. Pathogens. 2021. PMID: 34578208 Free PMC article. Review.
-
Genotypic and Phenotypic Diversity of Staphylococcus aureus Isolates from Cystic Fibrosis Patient Lung Infections and Their Interactions with Pseudomonas aeruginosa.mBio. 2020 Jun 23;11(3):e00735-20. doi: 10.1128/mBio.00735-20. mBio. 2020. PMID: 32576671 Free PMC article.
-
Regulation of airway fumarate by host and pathogen promotes Staphylococcus aureus pneumonia.Nat Commun. 2025 Aug 1;16(1):7050. doi: 10.1038/s41467-025-62453-y. Nat Commun. 2025. PMID: 40745169 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

