Phylogenomic Analysis of Extraintestinal Pathogenic Escherichia coli Sequence Type 1193, an Emerging Multidrug-Resistant Clonal Group
- PMID: 30348668
- PMCID: PMC6325179
- DOI: 10.1128/AAC.01913-18
Phylogenomic Analysis of Extraintestinal Pathogenic Escherichia coli Sequence Type 1193, an Emerging Multidrug-Resistant Clonal Group
Abstract
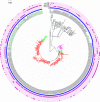
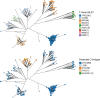
The fluoroquinolone-resistant sequence type 1193 (ST1193) of Escherichia coli, from the ST14 clonal complex (STc14) within phylogenetic group B2, has appeared recently as an important cause of extraintestinal disease in humans. Although this emerging lineage has been characterized to some extent using conventional methods, it has not been studied extensively at the genomic level. Here, we used whole-genome sequence analysis to compare 355 ST1193 isolates with 72 isolates from other STs within STc14. Using core genome phylogeny, the ST1193 isolates formed a tightly clustered clade with many genotypic similarities, unlike ST14 isolates. All ST1193 isolates possessed the same set of three chromosomal mutations conferring fluoroquinolone resistance, carried the fimH64 allele, and were lactose non-fermenting. Analysis revealed an evolutionary progression from K1 to K5 capsular types and acquisition of an F-type virulence plasmid, followed by changes in plasmid structure congruent with genome phylogeny. In contrast, the numerous identified antimicrobial resistance genes were distributed incongruently with the underlying phylogeny, suggesting frequent gain or loss of the corresponding resistance gene cassettes despite retention of the presumed carrier plasmids. Pangenome analysis revealed gains and losses of genetic loci occurring during the transition from ST14 to ST1193 and from the K1 to K5 capsular types. Using time-scaled phylogenetic analysis, we estimated that current ST1193 clades first emerged approximately 25 years ago. Overall, ST1193 appears to be a recently emerged clone in which both stepwise and mosaic evolution have contributed to epidemiologic success.
Keywords: ExPEC; ST1193; antimicrobial resistance; fluoroquinolone; plasmid.
Copyright © 2018 American Society for Microbiology.
Figures





References
-
- Colpan A, Johnston B, Porter S, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR, Allen BL, Baracco GJ, Bedimo R, Bessesen M, Bonomo RA, Brecher SM, Brown ST, Castellino L, Desai AS, Fernau F, Fisher MA, Fleckenstein J, Fleming CS, Fries NJ, Kan VL, Kauffman CA, Klutts S, Ohl M, Russo T, Swiatlo A, Swiatlo E. 2013. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin Infect Dis 57:1256–1265. doi: 10.1093/cid/cit503. - DOI - PMC - PubMed
-
- Tchesnokova V, Rechkina E, Larson L, Ferrier K, Weaver JL, Schroeder DW, She R, Butler-Wu SM, Aguero-Rosenfeld ME, Zerr D, Fang FC, Ralston J, Riddell K, Scholes D, Weissman S, Parker K, Spellberg B, Johnson JR, Sokurenko EV. 29 June 2018. Rapid and extensive expansion in the U.S. of a new multidrug-resistant Escherichia coli clonal group, sequence type ST1193. Clin Infect Dis doi: 10.1093/cid/ciy525. - DOI - PMC - PubMed
-
- Platell JL, Trott DJ, Johnson JR, Heisig P, Heisig A, Clabots CR, Johnston B, Cobbold RN. 2012. Prominence of an O75 clonal group (clonal complex 14) among non-st131 fluoroquinolone-resistant Escherichia coli causing extraintestinal infections in humans and dogs in Australia. Antimicrob Agents Chemother 56:3898–3904. doi: 10.1128/AAC.06120-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

