Ribosome biogenesis factor Ltv1 chaperones the assembly of the small subunit head
- PMID: 30348748
- PMCID: PMC6279377
- DOI: 10.1083/jcb.201804163
Ribosome biogenesis factor Ltv1 chaperones the assembly of the small subunit head
Abstract
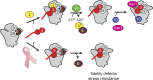
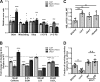
The correct assembly of ribosomes from ribosomal RNAs (rRNAs) and ribosomal proteins (RPs) is critical, as indicated by the diseases caused by RP haploinsufficiency and loss of RP stoichiometry in cancer cells. Nevertheless, how assembly of each RP is ensured remains poorly understood. We use yeast genetics, biochemistry, and structure probing to show that the assembly factor Ltv1 facilitates the incorporation of Rps3, Rps10, and Asc1/RACK1 into the small ribosomal subunit head. Ribosomes from Ltv1-deficient yeast have substoichiometric amounts of Rps10 and Asc1 and show defects in translational fidelity and ribosome-mediated RNA quality control. These defects provide a growth advantage under some conditions but sensitize the cells to oxidative stress. Intriguingly, relative to glioma cell lines, breast cancer cells have reduced levels of LTV1 and produce ribosomes lacking RPS3, RPS10, and RACK1. These data describe a mechanism to ensure RP assembly and demonstrate how cancer cells circumvent this mechanism to generate diverse ribosome populations that can promote survival under stress.
© 2018 Collins et al.
Figures






References
-
- Ajore R., Raiser D., McConkey M., Jöud M., Boidol B., Mar B., Saksena G., Weinstock D.M., Armstrong S., Ellis S.R., et al. 2017. Deletion of ribosomal protein genes is a common vulnerability in human cancer, especially in concert with TP53 mutations. EMBO Mol. Med. 9:498–507. 10.15252/emmm.201606660 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

