Identification and validation of a tumor-infiltrating Treg transcriptional signature conserved across species and tumor types
- PMID: 30348759
- PMCID: PMC6233093
- DOI: 10.1073/pnas.1810580115
Identification and validation of a tumor-infiltrating Treg transcriptional signature conserved across species and tumor types
Abstract
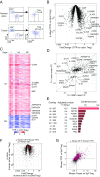
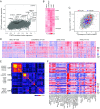
FoxP3+ T regulatory (Treg) cells are central elements of immunologic tolerance. They are abundant in many tumors, where they restrict potentially favorable antitumor responses. We used a three-pronged strategy to identify genes related to the presence and function of Tregs in the tumor microenvironment. Gene expression profiles were generated from tumor-infiltrating Tregs (TITRs) of both human and mouse tumors and were compared with those of Tregs of lymphoid organs or normal tissues from the same individuals. A computational deconvolution of whole-tumor datasets from the Cancer Genome Atlas (TCGA) was performed to identify transcripts specifically associated with Tregs across thousands of tumors from different stages and locations. We identified a set of TITR-differential transcripts with striking reproducibility between tumor types in mice, between mice and humans, and between different human patients spanning tumor stages. Many of the TITR-preferential transcripts were shared with "tissue Tregs" residing in nonlymphoid tissues, but a tumor-preferential segment could be identified. Many of these TITR signature transcripts were confirmed by mining of TCGA datasets, which also brought forth transcript modules likely representing the parenchymal attraction of, or response to, tumor Tregs. Importantly, the TITR signature included several genes encoding effective targets of tumor immunotherapy. A number of other targets were validated by CRISPR-based gene inactivation in mouse Tregs. These results confirm the validity of the signature, generating a wealth of leads for understanding the role of Tregs in tumor progression and identifying potential targets for cancer immunotherapy.
Keywords: T cell differentiation; immuno-oncology; immunotherapy.
Conflict of interest statement
Conflict of interest statement: C.B. co-authored a consortium position paper with Miriam Merad in 2017 that stems from the Human Cell Atlas, a large consortium to which they both belong; they did not collaborate directly on the paper. C.B. is a principal investigator in a bridge funding for a consortium (Immune Cell Atlas), of which Dr. Merad is the Program Director; Dr. Merad’s review of this paper was concluded before this award.
Figures




References
-
- Benoist C, Mathis D. In: Immune Tolerance. Mathis D, Rudensky A, editors. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2013. pp. 31–44.
-
- Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. - PubMed
-
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. - PubMed
-
- Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

