Biochemical and structural studies reveal differences and commonalities among cap-snatching endonucleases from segmented negative-strand RNA viruses
- PMID: 30348898
- PMCID: PMC6314124
- DOI: 10.1074/jbc.RA118.004373
Biochemical and structural studies reveal differences and commonalities among cap-snatching endonucleases from segmented negative-strand RNA viruses
Abstract
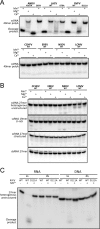
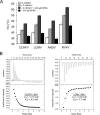
Viruses rely on many host cell processes, including the cellular transcription machinery. Segmented negative-strand RNA viruses (sNSV) in particular cannot synthesize the 5'-cap structure for their mRNA but cleave off cellular caps and use the resulting oligonucleotides as primers for their transcription. This cap-snatching mechanism, involving a viral cap-binding site and RNA endonuclease, is both virus-specific and essential for viral proliferation and therefore represents an attractive drug target. Here, we present biochemical and structural results on the putative cap-snatching endonuclease of Crimean-Congo hemorrhagic fever virus (CCHFV), a highly pathogenic bunyavirus belonging to the Nairoviridae family, and of two additional nairoviruses, Erve virus (EREV) and Nairobi sheep disease virus (NSDV). Our findings are presented in the context of other cap-snatching endonucleases, such as the enzymatically active endonuclease from Rift Valley fever virus (RVFV), from Arenaviridae and Bunyavirales, belonging to the His- and His+ endonucleases, respectively, according to the absence or presence of a metal ion-coordinating histidine in the active site. Mutational and metal-binding experiments revealed the presence of only acidic metal-coordinating residues in the active site of the CCHFV domain and a unique active-site conformation that was intermediate between those of His+ and His- endonucleases. On the basis of small-angle X-ray scattering (SAXS) and homology modeling results, we propose a protein topology for the CCHFV domain that, despite its larger size, has a structure overall similar to those of related endonucleases. These results suggest structural and functional conservation of the cap-snatching mechanism among sNSVs.
Keywords: cap-snatching; endonuclease; enzyme structure; metal ion–protein interaction; nairovirus; negative-strand RNA virus; recombinant protein expression; small-angle X-ray scattering (SAXS); structure-function; viral transcription.
© 2018 Holm et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

