Monoubiquitination of ASXLs controls the deubiquitinase activity of the tumor suppressor BAP1
- PMID: 30349006
- PMCID: PMC6197237
- DOI: 10.1038/s41467-018-06854-2
Monoubiquitination of ASXLs controls the deubiquitinase activity of the tumor suppressor BAP1
Abstract
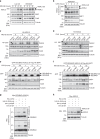
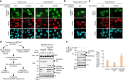
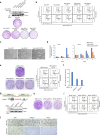
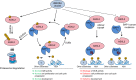
The tumor suppressor and deubiquitinase (DUB) BAP1 and its Drosophila ortholog Calypso assemble DUB complexes with the transcription regulators Additional sex combs-like (ASXL1, ASXL2, ASXL3) and Asx respectively. ASXLs and Asx use their DEUBiquitinase ADaptor (DEUBAD) domain to stimulate BAP1/Calypso DUB activity. Here we report that monoubiquitination of the DEUBAD is a general feature of ASXLs and Asx. BAP1 promotes DEUBAD monoubiquitination resulting in an increased stability of ASXL2, which in turn stimulates BAP1 DUB activity. ASXL2 monoubiquitination is directly catalyzed by UBE2E family of Ubiquitin-conjugating enzymes and regulates mammalian cell proliferation. Remarkably, Calypso also regulates Asx monoubiquitination and transgenic flies expressing monoubiquitination-defective Asx mutant exhibit developmental defects. Finally, the protein levels of ASXL2, BAP1 and UBE2E enzymes are highly correlated in mesothelioma tumors suggesting the importance of this signaling axis for tumor suppression. We propose that monoubiquitination orchestrates a molecular symbiosis relationship between ASXLs and BAP1.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

