Long non-coding RNA-dependent mechanism to regulate heme biosynthesis and erythrocyte development
- PMID: 30349036
- PMCID: PMC6197277
- DOI: 10.1038/s41467-018-06883-x
Long non-coding RNA-dependent mechanism to regulate heme biosynthesis and erythrocyte development
Abstract
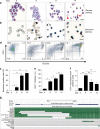
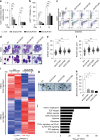
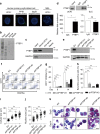
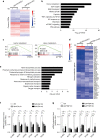
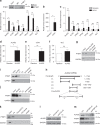
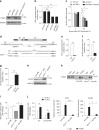
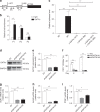
In addition to serving as a prosthetic group for enzymes and a hemoglobin structural component, heme is a crucial homeostatic regulator of erythroid cell development and function. While lncRNAs modulate diverse physiological and pathological cellular processes, their involvement in heme-dependent mechanisms is largely unexplored. In this study, we elucidated a lncRNA (UCA1)-mediated mechanism that regulates heme metabolism in human erythroid cells. We discovered that UCA1 expression is dynamically regulated during human erythroid maturation, with a maximal expression in proerythroblasts. UCA1 depletion predominantly impairs heme biosynthesis and arrests erythroid differentiation at the proerythroblast stage. Mechanistic analysis revealed that UCA1 physically interacts with the RNA-binding protein PTBP1, and UCA1 functions as an RNA scaffold to recruit PTBP1 to ALAS2 mRNA, which stabilizes ALAS2 mRNA. These results define a lncRNA-mediated posttranscriptional mechanism that provides a new dimension into how the fundamental heme biosynthetic process is regulated as a determinant of erythrocyte development.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
- 81870089/National Natural Science Foundation of China (National Science Foundation of China)/International
- 31471291/National Natural Science Foundation of China (National Science Foundation of China)/International
- 15JCYBJC54500/Natural Science Foundation of Tianjin City (Natural Science Foundation of Tianjin)/International
- R01 DK050107/DK/NIDDK NIH HHS/United States
- DK50107/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)/International
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

