Recruitment of LC3 to damaged Golgi apparatus
- PMID: 30349077
- PMCID: PMC6748261
- DOI: 10.1038/s41418-018-0221-5
Recruitment of LC3 to damaged Golgi apparatus
Abstract
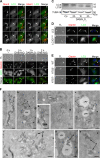
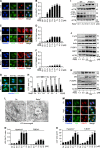
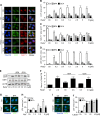
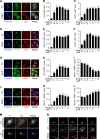
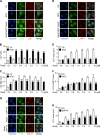
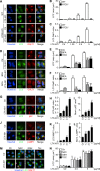
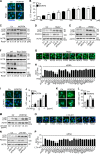
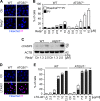
LC3 is a protein that can associate with autophagosomes, autolysosomes, and phagosomes. Here, we show that LC3 can also redistribute toward the damaged Golgi apparatus where it clusters with SQSTM1/p62 and lysosomes. This organelle-specific relocation, which did not involve the generation of double-membraned autophagosomes, could be observed after Golgi damage was induced by various strategies, namely (i) laser-induced localized cellular damage, (ii) local expression of peroxidase and exposure to peroxide and diaminobenzidine, (iii) treatment with the Golgi-tropic photosensitizer redaporfin and light, (iv) or exposure to the Golgi-tropic anticancer peptidomimetic LTX-401. Mechanistic exploration led to the conclusion that both reactive oxygen species-dependent and -independent Golgi damage induces a similar phenotype that depended on ATG5 yet did not depend on phosphatidylinositol-3-kinase catalytic subunit type 3 and Beclin-1. Interestingly, knockout of ATG5 sensitized cells to Golgi damage-induced cell death, suggesting that the pathway culminating in the relocation of LC3 to the damaged Golgi may have a cytoprotective function.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

