Evaluation of plant contamination in metabarcoding diet analysis of a herbivore
- PMID: 30349088
- PMCID: PMC6197254
- DOI: 10.1038/s41598-018-32845-w
Evaluation of plant contamination in metabarcoding diet analysis of a herbivore
Abstract
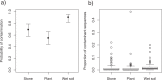
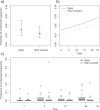
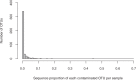
Fecal DNA metabarcoding is currently used in various fields of ecology to determine animal diets. Contamination of non-food DNA from complex field environments is a considerable challenge to the reliability of this method but has rarely been quantified. We evaluated plant DNA contamination by sequencing the chloroplast trnL P6 loop region from food-controlled geese feces. The average percentage of contaminant sequences per sample was 1.86%. According to the results of generalized linear models, the probability of contamination was highest in samples placed in wet soil. The proportion of contaminant sequences was lowest at the earliest sampling point and was slightly higher in samples placed in open conditions. Exclusion of rare OTUs (operational taxonomic units) was effective for obtaining reliable dietary data from the obtained sequences, and a 1% cutoff reduced the percentage of contaminated samples to less than 30%. However, appropriate interpretation of the barcoding results considering inevitable contamination is an important issue to address. We suggest the following procedures for fecal sampling and sequence data treatment to increase the reliability of DNA metabarcoding diet analyses: (i) Collect samples as soon as possible after deposition, (ii) avoid samples from deposits on wet soil, and (iii) exclude rare OTUs from diet composition estimations.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

