Long noncoding RNA MALAT1 suppresses breast cancer metastasis
- PMID: 30349115
- PMCID: PMC6265076
- DOI: 10.1038/s41588-018-0252-3
Long noncoding RNA MALAT1 suppresses breast cancer metastasis
Abstract
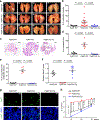
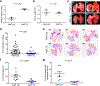
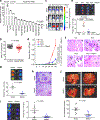
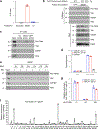
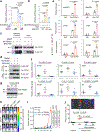
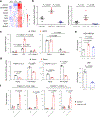
MALAT1 has previously been described as a metastasis-promoting long noncoding RNA (lncRNA). We show here, however, that targeted inactivation of the Malat1 gene in a transgenic mouse model of breast cancer, without altering the expression of its adjacent genes, promotes lung metastasis, and that this phenotype can be reversed by genetic add-back of Malat1. Similarly, knockout of MALAT1 in human breast cancer cells induces their metastatic ability, which is reversed by re-expression of Malat1. Conversely, overexpression of Malat1 suppresses breast cancer metastasis in transgenic, xenograft, and syngeneic models. Mechanistically, the MALAT1 lncRNA binds and inactivates the prometastatic transcription factor TEAD, preventing TEAD from associating with its co-activator YAP and target gene promoters. Moreover, MALAT1 levels inversely correlate with breast cancer progression and metastatic ability. These findings demonstrate that MALAT1 is a metastasis-suppressing lncRNA rather than a metastasis promoter in breast cancer, calling for rectification of the model for this highly abundant and conserved lncRNA.
Conflict of interest statement
Competing interests
The authors declare no completing interests.
Figures
Comment in
-
MALAT1 long non-coding RNA and breast cancer.RNA Biol. 2019 Jun;16(6):860-863. doi: 10.1080/15476286.2019.1592072. Epub 2019 Mar 22. RNA Biol. 2019. PMID: 30874469 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

