An atlas of genetic associations in UK Biobank
- PMID: 30349118
- PMCID: PMC6707814
- DOI: 10.1038/s41588-018-0248-z
An atlas of genetic associations in UK Biobank
Abstract
Genome-wide association studies (GWAS) have identified many loci contributing to variation in complex traits, yet the majority of loci that contribute to the heritability of complex traits remain elusive. Large study populations with sufficient statistical power are required to detect the small effect sizes of the yet unidentified genetic variants. However, the analysis of huge cohorts, like UK Biobank, is challenging. Here, we present an atlas of genetic associations for 118 non-binary and 660 binary traits of 452,264 UK Biobank participants of European ancestry. Results are compiled in a publicly accessible database that allows querying genome-wide association results for 9,113,133 genetic variants, as well as downloading GWAS summary statistics for over 30 million imputed genetic variants (>23 billion phenotype-genotype pairs). Our atlas of associations (GeneATLAS, http://geneatlas.roslin.ed.ac.uk ) will help researchers to query UK Biobank results in an easy and uniform way without the need to incur high computational costs.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
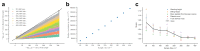
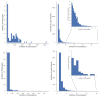
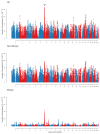


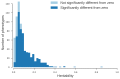

References
-
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. Longman; 1996.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

