Mutational analysis of the conserved carboxylates of anion channelrhodopsin-2 (ACR2) expressed in Escherichia coli and their roles in anion transport
- PMID: 30349802
- PMCID: PMC6194965
- DOI: 10.2142/biophysico.15.0_179
Mutational analysis of the conserved carboxylates of anion channelrhodopsin-2 (ACR2) expressed in Escherichia coli and their roles in anion transport
Abstract
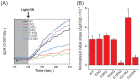
Anion channelrhodopsin-2 (ACR2), a light-gated channel recently identified from the cryptophyte alga Guillardia theta, exhibits anion channel activity with exclusive selectivity. In addition to its novel function, ACR2 has become a focus of interest as a powerful tool for optogenetics. Here we combined experimental and computational approaches to investigate the roles of conserved carboxylates on the anion transport activity of ACR2 in Escherichia coli membrane. First, we replaced six conserved carboxylates with a neutral residue (i.e. E9Q, E56Q, E64Q, E159Q, E219Q and D230N), and measured anion transport activity using E. coli expression system. E159Q and D230N exhibited significantly lower anion transport activity compared with wild-type ACR2 (1/12~1/3.4), which suggests that E159 and D230 play important roles in the anion transport. Second, to explain its molecular aspects, we constructed a homology model of ACR2 based on the crystal structure of a cation channelrhodopsin (ChR). The model structure showed a cavity formed by four transmembrane helices (TM1, TM2, TM3 and TM7) similar to ChRs, as a putative anion conducting pathway. Although E159 is not located in the putative pathway, the model structure showed hydrogen bonds between E159 and R129 with a water molecule. D230 is located in the pathway near the protonated Schiff base (PSB) of the chromophore retinal, which suggests that there is an interaction between D230 and the PSB. Thus, we demonstrated the functional importance and the hypothetical roles of two conserved carboxylates, E159 and D230, in the anion transport activity of ACR2 in E. coli membrane.
Keywords: anion channel; ion transport; microbial rhodopsin; retinal.
Conflict of interest statement
Conflict of Interest There are no conflicts of interest to declare. The authors declare no competing financial interests.
Figures





References
-
- Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nature New Biol. 1971;233:149–152. - PubMed
LinkOut - more resources
Full Text Sources
