Variability of Betweenness Centrality and Its Effect on Identifying Essential Genes
- PMID: 30350013
- PMCID: PMC6476710
- DOI: 10.1007/s11538-018-0526-z
Variability of Betweenness Centrality and Its Effect on Identifying Essential Genes
Abstract
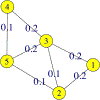
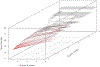
This paper begins to build a theoretical framework that would enable the pharmaceutical industry to use network complexity measures as a way to identify drug targets. The variability of a betweenness measure for a network node is examined through different methods of network perturbation. Our results indicate a robustness of betweenness centrality in the identification of target genes.
Keywords: Betweenness centrality; Differential expression; Network complexity measure.
Figures
References
-
- Han J-DJ, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, et al., “Evidence for dynamically organized modularity in the yeast protein–protein interaction network,” Nature, vol. 430, no. 6995, p. 88, 2004. - PubMed
-
- Jeong H, Mason SP, Barabási A-L, and Oltvai ZN, “Lethality and centrality in protein networks,” Nature, vol. 411, no. 6833, p. 41, 2001. - PubMed
-
- Freeman LC, “A set of measures of centrality based on betweenness,” Sociometry, pp. 35–41, 1977.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

