PLENTY, a hydroxyproline O-arabinosyltransferase, negatively regulates root nodule symbiosis in Lotus japonicus
- PMID: 30351431
- PMCID: PMC6322572
- DOI: 10.1093/jxb/ery364
PLENTY, a hydroxyproline O-arabinosyltransferase, negatively regulates root nodule symbiosis in Lotus japonicus
Abstract
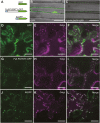
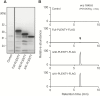
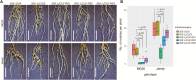
Legumes can survive in nitrogen-deficient environments by forming root-nodule symbioses with rhizobial bacteria; however, forming nodules consumes energy, and nodule numbers must thus be strictly controlled. Previous studies identified major negative regulators of nodulation in Lotus japonicus, including the small peptides CLAVATA3/ESR (CLE)-RELATED-ROOT SIGNAL1 (CLE-RS1), CLE-RS2, and CLE-RS3, and their putative major receptor HYPERNODULATION AND ABERRANT ROOT FORMATION1 (HAR1). CLE-RS2 is known to be expressed in rhizobia-inoculated roots, and is predicted to be post-translationally arabinosylated, a modification essential for its activity. Moreover, all three CLE-RSs suppress nodulation in a HAR1-dependent manner. Here, we identified PLENTY as a gene responsible for the previously isolated hypernodulation mutant plenty. PLENTY encoded a hydroxyproline O-arabinosyltransferase orthologous to ROOT DETERMINED NODULATION1 in Medicago truncatula. PLENTY was localized to the Golgi, and an in vitro analysis of the recombinant protein demonstrated its arabinosylation activity, indicating that CLE-RS1/2/3 may be substrates for PLENTY. The constitutive expression experiments showed that CLE-RS3 was the major candidate substrate for PLENTY, suggesting the substrate preference of PLENTY for individual CLE-RS peptides. Furthermore, a genetic analysis of the plenty har1 double mutant indicated the existence of another PLENTY-dependent and HAR1-independent pathway negatively regulating nodulation.
Figures


Similar articles
-
CLE-HAR1 Systemic Signaling and NIN-Mediated Local Signaling Suppress the Increased Rhizobial Infection in the daphne Mutant of Lotus japonicus.Mol Plant Microbe Interact. 2020 Feb;33(2):320-327. doi: 10.1094/MPMI-08-19-0223-R. Epub 2019 Dec 27. Mol Plant Microbe Interact. 2020. PMID: 31880983
-
Expression of the CLE-RS3 gene suppresses root nodulation in Lotus japonicus.J Plant Res. 2016 Sep;129(5):909-919. doi: 10.1007/s10265-016-0842-z. Epub 2016 Jun 13. J Plant Res. 2016. PMID: 27294965
-
Nodule Inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production.Proc Natl Acad Sci U S A. 2014 Oct 7;111(40):14607-12. doi: 10.1073/pnas.1412716111. Epub 2014 Sep 22. Proc Natl Acad Sci U S A. 2014. PMID: 25246578 Free PMC article.
-
Leguminous plants: inventors of root nodules to accommodate symbiotic bacteria.Int Rev Cell Mol Biol. 2015;316:111-58. doi: 10.1016/bs.ircmb.2015.01.004. Epub 2015 Feb 20. Int Rev Cell Mol Biol. 2015. PMID: 25805123 Review.
-
Nitrate transporters: an overview in legumes.Planta. 2017 Oct;246(4):585-595. doi: 10.1007/s00425-017-2724-6. Epub 2017 Jun 26. Planta. 2017. PMID: 28653185 Review.
Cited by
-
Progress in the Self-Regulation System in Legume Nodule Development-AON (Autoregulation of Nodulation).Int J Mol Sci. 2022 Jun 15;23(12):6676. doi: 10.3390/ijms23126676. Int J Mol Sci. 2022. PMID: 35743118 Free PMC article. Review.
-
The impact of the rhizobia-legume symbiosis on host root system architecture.J Exp Bot. 2020 Jun 26;71(13):3902-3921. doi: 10.1093/jxb/eraa198. J Exp Bot. 2020. PMID: 32337556 Free PMC article. Review.
-
Plants Recruit Peptides and Micro RNAs to Regulate Nutrient Acquisition from Soil and Symbiosis.Plants (Basel). 2023 Jan 2;12(1):187. doi: 10.3390/plants12010187. Plants (Basel). 2023. PMID: 36616316 Free PMC article. Review.
-
Shoot Extracts from Two Low Nodulation Mutants Significantly Reduce Nodule Number in Pea.Plants (Basel). 2020 Nov 6;9(11):1505. doi: 10.3390/plants9111505. Plants (Basel). 2020. PMID: 33172149 Free PMC article.
-
Plant Protein O-Arabinosylation.Front Plant Sci. 2021 Mar 18;12:645219. doi: 10.3389/fpls.2021.645219. eCollection 2021. Front Plant Sci. 2021. PMID: 33815452 Free PMC article. Review.
References
-
- Caetano-Anollés G, Gresshoff PM. 1990. Early induction of feedback regulatory responses governing nodulation in soybean. Plant Science 71, 69–81.
-
- Corcilius L, Hastwell AH, Zhang M, Williams J, Mackay JP, Gresshoff PM, Ferguson BJ, Payne RJ. 2017. Arabinosylation modulates the growth-regulating activity of the peptide hormone CLE40a from soybean. Cell Chemical Biology 24, 1347–1355. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

