DNA replication and repair kinetics of Alu, LINE-1 and satellite III genomic repetitive elements
- PMID: 30352618
- PMCID: PMC6198450
- DOI: 10.1186/s13072-018-0226-9
DNA replication and repair kinetics of Alu, LINE-1 and satellite III genomic repetitive elements
Abstract
Background: Preservation of genome integrity by complete, error-free DNA duplication prior to cell division and by correct DNA damage repair is paramount for the development and maintenance of an organism. This holds true not only for protein-encoding genes, but also it applies to repetitive DNA elements, which make up more than half of the human genome. Here, we focused on the replication and repair kinetics of interspersed and tandem repetitive DNA elements.
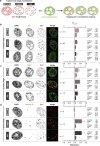
Results: We integrated genomic population level data with a single cell immunofluorescence in situ hybridization approach to simultaneously label replication/repair and repetitive DNA elements. We found that: (1) the euchromatic Alu element was replicated during early S-phase; (2) LINE-1, which is associated with AT-rich genomic regions, was replicated throughout S-phase, with the majority being replicated according to their particular histone marks; (3) satellite III, which constitutes pericentromeric heterochromatin, was replicated exclusively during the mid-to-late S-phase. As for the DNA double-strand break repair process, we observed that Alu elements followed the global genome repair kinetics, while LINE-1 elements repaired at a slower rate. Finally, satellite III repeats were repaired at later time points.
Conclusions: We conclude that the histone modifications in the specific repeat element predominantly determine its replication and repair timing. Thus, Alu elements, which are characterized by euchromatic chromatin features, are repaired and replicated the earliest, followed by LINE-1 elements, including more variegated eu/heterochromatic features and, lastly, satellite tandem repeats, which are homogeneously characterized by heterochromatic features and extend over megabase-long genomic regions. Altogether, this work reemphasizes the need for complementary approaches to achieve an integrated and comprehensive investigation of genomic processes.
Keywords: Alu; ChIP-Seq; DNA repair; DNA repetitive elements; DNA replication; Genome-wide analysis; Immuno-FISH; LINE; Phosphorylated H2AX; Satellites.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

