Targeted genotyping of variable number tandem repeats with adVNTR
- PMID: 30352806
- PMCID: PMC6211647
- DOI: 10.1101/gr.235119.118
Targeted genotyping of variable number tandem repeats with adVNTR
Abstract
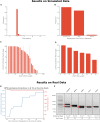
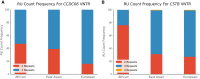
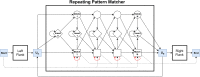
Whole-genome sequencing is increasingly used to identify Mendelian variants in clinical pipelines. These pipelines focus on single-nucleotide variants (SNVs) and also structural variants, while ignoring more complex repeat sequence variants. Here, we consider the problem of genotyping Variable Number Tandem Repeats (VNTRs), composed of inexact tandem duplications of short (6-100 bp) repeating units. VNTRs span 3% of the human genome, are frequently present in coding regions, and have been implicated in multiple Mendelian disorders. Although existing tools recognize VNTR carrying sequence, genotyping VNTRs (determining repeat unit count and sequence variation) from whole-genome sequencing reads remains challenging. We describe a method, adVNTR, that uses hidden Markov models to model each VNTR, count repeat units, and detect sequence variation. adVNTR models can be developed for short-read (Illumina) and single-molecule (Pacific Biosciences [PacBio]) whole-genome and whole-exome sequencing, and show good results on multiple simulated and real data sets.
© 2018 Bakhtiari et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. - PubMed
-
- Benedetti F, Dallaspezia S, Colombo C, Pirovano A, Marino E, Smeraldi E. 2008. A length polymorphism in the circadian clock gene Per3 influences age at onset of bipolar disorder. Neurosci Lett 445: 184–187. - PubMed
-
- Berlin K, Koren S, Chin CS, Drake JP, Landolin JM, Phillippy AM. 2015. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol 33: 623–630. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
