The mechanism of resistance to favipiravir in influenza
- PMID: 30352857
- PMCID: PMC6233120
- DOI: 10.1073/pnas.1811345115
The mechanism of resistance to favipiravir in influenza
Abstract
Favipiravir is a broad-spectrum antiviral that has shown promise in treatment of influenza virus infections. While emergence of resistance has been observed for many antiinfluenza drugs, to date, clinical trials and laboratory studies of favipiravir have not yielded resistant viruses. Here we show evolution of resistance to favipiravir in the pandemic H1N1 influenza A virus in a laboratory setting. We found that two mutations were required for robust resistance to favipiravir. We demonstrate that a K229R mutation in motif F of the PB1 subunit of the influenza virus RNA-dependent RNA polymerase (RdRP) confers resistance to favipiravir in vitro and in cell culture. This mutation has a cost to viral fitness, but fitness can be restored by a P653L mutation in the PA subunit of the polymerase. K229R also conferred favipiravir resistance to RNA polymerases of other influenza A virus strains, and its location within a highly conserved structural feature of the RdRP suggests that other RNA viruses might also acquire resistance through mutations in motif F. The mutations identified here could be used to screen influenza virus-infected patients treated with favipiravir for the emergence of resistance.
Keywords: antiviral; influenza; polymerase; resistance; virus.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



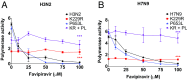


References
-
- WHO 2018 Influenza (Seasonal) Factsheet. Available at www.who.int/mediacentre/factsheets/fs211/en. Accessed February 22, 2018.
-
- Hurt AC. The epidemiology and spread of drug resistant human influenza viruses. Curr Opin Virol. 2014;8:22–29. - PubMed
-
- Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA. 2006;295:891–894. - PubMed
-
- Weinstock DM, Zuccotti G. The evolution of influenza resistance and treatment. JAMA. 2009;301:1066–1069. - PubMed
-
- Weinstock DM, Gubareva LV, Zuccotti G. Prolonged shedding of multidrug-resistant influenza A virus in an immunocompromised patient. N Engl J Med. 2003;348:867–868. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

