Achieving a multi-strain symbiosis: strain behavior and infection dynamics
- PMID: 30353039
- PMCID: PMC6461934
- DOI: 10.1038/s41396-018-0305-8
Achieving a multi-strain symbiosis: strain behavior and infection dynamics
Abstract
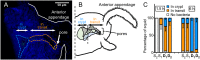
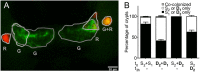
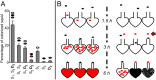
Strain diversity, while now recognized as a key driver underlying partner dynamics in symbioses, is usually difficult to experimentally manipulate and image in hosts with complex microbiota. To address this problem, we have used the luminous marine bacterium Vibrio fischeri, which establishes a symbiosis within the crypts of the nascent light organ of the squid Euprymna scolopes. Competition assays in newly hatched juvenile squid have shown that symbiotic V. fischeri are either niche-sharing "S strains", which share the light organ when co-inoculated with other S strains, or niche-dominant "D strains", which are typically found alone in the light organ after a co-colonization. To understand this D strain advantage, we determined the minimum time that different V. fischeri strains needed to initiate colonization and used confocal microscopy to localize the symbionts along their infection track. Further, we determined whether symbiont-induced host morphogenic events also occurred earlier during a D strain colonization. We conclude that D strains colonized more quickly than S strains. Nevertheless, light-organ populations in field-caught adult squid often contain both D and S strains. We determined experimentally that this symbiont population heterogeneity might be achieved in nature by a serial encounter of different strains in the environment.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Grants and funding
- GM099507/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)/International
- R01 AI050661/AI/NIAID NIH HHS/United States
- OD011024/U.S. Department of Health & Human Services | NIH | NIH Office of the Director (OD)/International
- R01 RR012294/RR/NCRR NIH HHS/United States
- R01 GM099507/GM/NIGMS NIH HHS/United States

