c-Fos induces chondrogenic tumor formation in immortalized human mesenchymal progenitor cells
- PMID: 30353072
- PMCID: PMC6199246
- DOI: 10.1038/s41598-018-33689-0
c-Fos induces chondrogenic tumor formation in immortalized human mesenchymal progenitor cells
Abstract
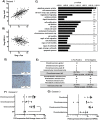
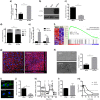
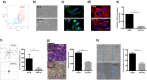
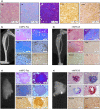
Mesenchymal progenitor cells (MPCs) have been hypothesized as cells of origin for sarcomas, and c-Fos transcription factor has been showed to act as an oncogene in bone tumors. In this study, we show c-Fos is present in most sarcomas with chondral phenotype, while multiple other genes are related to c-Fos expression pattern. To further define the role of c-Fos in sarcomagenesis, we expressed it in primary human MPCs (hMPCs), immortalized hMPCs and transformed murine MPCs (mMPCs). In immortalized hMPCs, c-Fos expression generated morphological changes, reduced mobility capacity and impaired adipogenic- and osteogenic-differentiation potentials. Remarkably, immortalized hMPCs or mMPCs expressing c-Fos generated tumors harboring a chondrogenic phenotype and morphology. Thus, here we show that c-Fos protein has a key role in sarcomas and that c-Fos expression in immortalized MPCs yields cell transformation and chondrogenic tumor formation.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

