Ordering up gene expression by slowing down transcription factor binding kinetics
- PMID: 30353359
- PMCID: PMC6421095
- DOI: 10.1007/s00294-018-0896-7
Ordering up gene expression by slowing down transcription factor binding kinetics
Abstract
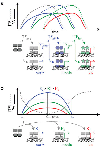
Efficient regulation of a complex genetic response requires that the gene products, which catalyze the response, be synthesized in a temporally ordered manner to match the sequential nature of the reaction pathway they act upon. Transcription regulation networks coordinate this aspect of cellular control by modulating transcription factor (TF) concentrations through time. The effect a TF has on the timing of gene expression is often modeled assuming that the TF-promoter binding reaction is in thermodynamic equilibrium with changes in TF concentration over time; however, non-equilibrium dynamics resulting from relatively slow TF-binding kinetics can result in different network behavior. Here, I highlight a recent study of the bacterial SOS response, where a single TF regulates multiple target promoters, to show how a disequilibrium of TF binding at promoters results in a more complex behavior, enabling a larger temporal separation of promoter activities that depends not only upon slow TF binding kinetics at promoters, but also on the magnitude of the response stimulus. I also discuss the dependence of network behavior on specific TF regulatory mechanisms and the implications non-equilibrium dynamics have for stochastic gene expression.
Keywords: DNA damage; Kinetics; LexA; Promoter; SOS response; Transcription factor.
Conflict of interest statement
Conflict of Interest: The author declares that he has no conflict of interest.
Figures

References
-
- Butala M, Klose D, Hodnik V, Rems A, Podlesek Z, Klare JP, Anderluh G, Busby SJ, Steinhoff HJ, Zgur-Bertok D (2011) Interconversion between bound and free conformations of LexA orchestrates the bacterial SOS response. Nucleic Acids Res. 39:6546–6557. doi: 10.1093/nar/gkr265 [doi] - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

