Role of DNA De Novo (De)Methylation in the Kidney in Salt-Induced Hypertension
- PMID: 30354815
- PMCID: PMC6314686
- DOI: 10.1161/HYPERTENSIONAHA.118.11650
Role of DNA De Novo (De)Methylation in the Kidney in Salt-Induced Hypertension
Abstract
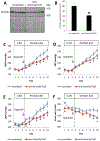
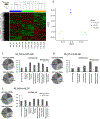
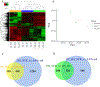
Numerous adult diseases involving tissues consisting primarily of nondividing cells are associated with changes in DNA methylation. It suggests a pathophysiological role for de novo methylation or demethylation of DNA, which is catalyzed by DNA methyltransferase 3 and ten-eleven translocases. However, the contribution of DNA de novo (de)methylation to these diseases remains almost completely unproven. Broad changes in DNA methylation occurred within days in the renal outer medulla of Dahl SS rats fed a high-salt diet, a classic model of hypertension. Intrarenal administration of anti-DNA methyltransferase 3a/ten-eleven translocase 3 GapmeRs attenuated high salt-induced hypertension in SS rats. The high-salt diet induced differential expression of 1712 genes in the renal outer medulla. Remarkably, the differential expression of 76% of these genes was prevented by anti-DNA methyltransferase 3a/ten-eleven translocase 3 GapmeRs. The genes differentially expressed in response to the GapmeRs were involved in the regulation of metabolism and inflammation and were significantly enriched for genes showing differential methylation in response to the GapmeRs. These data indicate a significant role of DNA de novo (de)methylation in the kidney in the development of hypertension in SS rats. The findings should help to shift the paradigm of DNA methylation research in diseases involving nondividing cells from correlative analysis to functional and mechanistic studies.
Keywords: DNA methylation; diet; genomics; hypertension; kidney.
Figures



Comment in
-
De Novo DNA (de)Methylation in the Kidney.Hypertension. 2018 Nov;72(5):1084-1086. doi: 10.1161/HYPERTENSIONAHA.118.11755. Hypertension. 2018. PMID: 30354833 Free PMC article. No abstract available.
References
-
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–57. - PubMed
-
- Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. 2017; 18(9):517–534. - PubMed
-
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14(3):204–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

