Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection
- PMID: 30355154
- PMCID: PMC6345613
- DOI: 10.1146/annurev-pathmechdis-012418-012751
Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection
Abstract
Nearly all infectious agents contain DNA or RNA genomes, making sequencing an attractive approach for pathogen detection. The cost of high-throughput or next-generation sequencing has been reduced by several orders of magnitude since its advent in 2004, and it has emerged as an enabling technological platform for the detection and taxonomic characterization of microorganisms in clinical samples from patients. This review focuses on the application of untargeted metagenomic next-generation sequencing to the clinical diagnosis of infectious diseases, particularly in areas in which conventional diagnostic approaches have limitations. The review covers ( a) next-generation sequencing technologies and common platforms, ( b) next-generation sequencing assay workflows in the clinical microbiology laboratory, ( c) bioinformatics analysis of metagenomic next-generation sequencing data, ( d) validation and use of metagenomic next-generation sequencing for diagnosing infectious diseases, and ( e) significant case reports and studies in this area. Next-generation sequencing is a new technology that has the promise to enhance our ability to diagnose, interrogate, and track infectious diseases.
Keywords: clinical diagnostics; infectious disease; metagenomics; nanopore sequencing; next-generation sequencing; pathogen detection.
Figures
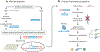
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

