Isolation and characterization of malaria PfHRP2 specific VNAR antibody fragments from immunized shark phage display library
- PMID: 30355309
- PMCID: PMC6201582
- DOI: 10.1186/s12936-018-2531-y
Isolation and characterization of malaria PfHRP2 specific VNAR antibody fragments from immunized shark phage display library
Abstract
Background: Malaria rapid diagnostic tests (RDTs) represent an important antibody based immunoassay platform. Unfortunately, conventional monoclonal antibodies are subject to degradation shortening shelf lives of RDTs. The variable region of the receptor (VNAR) from shark has a potential as alternative to monoclonal antibodies in RDTs due to high thermal stability.
Methods: In this study, new binders derived from shark VNAR domains library were investigated. Following immunization of a wobbegong shark (Orectolobus ornatus) with three recombinant malaria biomarker proteins (PfHRP2, PfpLDH and Pvaldolase), a single domain antibody (sdAb) library was constructed from splenocytes. Target-specific VNAR phage were isolated by panning. One specific clone was selected for expression in Escherichia coli expression system, and study of binding reactivity undertaken.
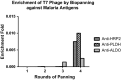
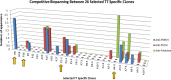
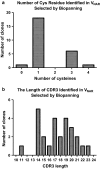
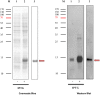
Results: The primary VNAR domain library possessed a titre of 1.16 × 106 pfu/mL. DNA sequence analysis showed 82.5% of isolated fragments appearing to contain an in-frame sequence. After multiple rounds of biopanning, a highly dominant clone specific to PfHRP2 was identified and selected for protein production in an E. coli expression system. Biological characterization showed the recombinant protein expressed in periplasmic has better detection sensitivity than that of cytoplasmic proteins. Assays of binding activity indicated that its reactivity was inferior to the positive control mAb C1-13.
Conclusions: Target-specific bacteriophage VNARs were successfully isolated after a series of immunization, demonstrating that phage display technology is a useful tool for selection of antigen binders. Generation of new binding reagents such as VNAR antibodies that specifically recognize the malaria biomarkers represents an appealing approach to improve the performance of RDTs.
Figures





Similar articles
-
Production and characterization of specific monoclonal antibodies binding the Plasmodium falciparum diagnostic biomarker, histidine-rich protein 2.Malar J. 2014 Jul 18;13:277. doi: 10.1186/1475-2875-13-277. Malar J. 2014. PMID: 25037150 Free PMC article.
-
Exploring shark VNAR antibody against infectious diseases using phage display technology.Fish Shellfish Immunol. 2023 Sep;140:108986. doi: 10.1016/j.fsi.2023.108986. Epub 2023 Aug 2. Fish Shellfish Immunol. 2023. PMID: 37541634 Review.
-
Cytoplasmic and periplasmic expression of recombinant shark VNAR antibody in Escherichia coli.Prep Biochem Biotechnol. 2019;49(4):315-327. doi: 10.1080/10826068.2019.1566145. Epub 2019 Feb 15. Prep Biochem Biotechnol. 2019. PMID: 30767708
-
Construction and next-generation sequencing analysis of a large phage-displayed VNAR single-domain antibody library from six naïve nurse sharks.Antib Ther. 2019 Jan;2(1):1-11. doi: 10.1093/abt/tby011. Epub 2018 Nov 7. Antib Ther. 2019. PMID: 30627698 Free PMC article.
-
Shark VNAR phage display libraries: An alternative source for therapeutic and diagnostic recombinant antibody fragments.Fish Shellfish Immunol. 2023 Jul;138:108808. doi: 10.1016/j.fsi.2023.108808. Epub 2023 May 9. Fish Shellfish Immunol. 2023. PMID: 37169114 Review.
Cited by
-
Progress on Phage Display Technology: Tailoring Antibodies for Cancer Immunotherapy.Viruses. 2023 Sep 9;15(9):1903. doi: 10.3390/v15091903. Viruses. 2023. PMID: 37766309 Free PMC article. Review.
-
Advances in the Production and Batch Reformatting of Phage Antibody Libraries.Mol Biotechnol. 2019 Nov;61(11):801-815. doi: 10.1007/s12033-019-00207-0. Mol Biotechnol. 2019. PMID: 31468301 Free PMC article. Review.
-
Chimeric Peptides from Californiconus californicus and Heterodontus francisci with Antigen-Binding Capacity: A Conotoxin Scaffold to Create Non-Natural Antibodies (NoNaBodies).Toxins (Basel). 2023 Apr 4;15(4):269. doi: 10.3390/toxins15040269. Toxins (Basel). 2023. PMID: 37104207 Free PMC article.
-
Shark IgNAR: The Next Broad Application Antibody in Clinical Diagnoses and Tumor Therapies?Mar Drugs. 2023 Sep 16;21(9):496. doi: 10.3390/md21090496. Mar Drugs. 2023. PMID: 37755109 Free PMC article. Review.
-
Diagnostic and therapeutic potential of shark variable new antigen receptor (VNAR) single domain antibody.Int J Biol Macromol. 2020 Mar 15;147:369-375. doi: 10.1016/j.ijbiomac.2020.01.039. Epub 2020 Jan 10. Int J Biol Macromol. 2020. PMID: 31926922 Free PMC article. Review.
References
-
- WHO . Malaria rapid diagnostic test performance. Geneva: World Health Organization; 2012.
-
- Kyabayinze DJ, Zongo I, Cunningham J, Gatton M, Angutoko P, Ategeka J, et al. HRP2 and pLDH-based rapid diagnostic tests, expert microscopy, and PCR for detection of malaria infection during pregnancy and at delivery in areas of varied transmission: a prospective cohort study in Burkina Faso and Uganda. PLoS One. 2016;11:e0156954. doi: 10.1371/journal.pone.0156954. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

