Efficient transposon mutagenesis mediated by an IPTG-controlled conditional suicide plasmid
- PMID: 30355324
- PMCID: PMC6201506
- DOI: 10.1186/s12866-018-1319-0
Efficient transposon mutagenesis mediated by an IPTG-controlled conditional suicide plasmid
Abstract
Background: Transposon mutagenesis is highly valuable for bacterial genetic and genomic studies. The transposons are usually delivered into host cells through conjugation or electroporation of a suicide plasmid. However, many bacterial species cannot be efficiently conjugated or transformed for transposon saturation mutagenesis. For this reason, temperature-sensitive (ts) plasmids have also been developed for transposon mutagenesis, but prolonged incubation at high temperatures to induce ts plasmid loss can be harmful to the hosts and lead to enrichment of mutants with adaptive genetic changes. In addition, the ts phenotype of a plasmid is often strain- or species-specific, as it may become non-ts or suicidal in different bacterial species.
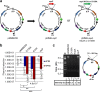
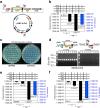
Results: We have engineered several conditional suicide plasmids that have a broad host range and whose loss is IPTG-controlled. One construct, which has the highest stability in the absence of IPTG induction, was then used as a curable vector to deliver hyperactive miniTn5 transposons for insertional mutagenesis. Our analyses show that these new tools can be used for efficient and regulatable transposon mutagenesis in Escherichia coli, Acinetobacter baylyi and Pseudomonas aeruginosa. In P. aeruginosa PAO1, we have used this method to generate a Tn5 insertion library with an estimated diversity of ~ 108, which is ~ 2 logs larger than the best transposon insertional library of PAO1 and related Pseudomonas strains previously reported.
Conclusion: We have developed a number of IPTG-controlled conditional suicide plasmids. By exploiting one of them for transposon delivery, a highly efficient and broadly useful mutagenesis system has been developed. As the assay condition is mild, we believe that our methodology will have broad applications in microbiology research.
Keywords: Acinetobacter; Conditional suicide plasmid; Escherichia coli; IPTG; Pseudomonas; Transposon mutagenesis.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements.J Bacteriol. 1996 Jul;178(14):4166-75. doi: 10.1128/jb.178.14.4166-4175.1996. J Bacteriol. 1996. PMID: 8763945 Free PMC article.
-
Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans.J Bacteriol. 1999 Dec;181(23):7298-307. doi: 10.1128/JB.181.23.7298-7307.1999. J Bacteriol. 1999. PMID: 10572134 Free PMC article.
-
Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria.J Bacteriol. 1990 Nov;172(11):6557-67. doi: 10.1128/jb.172.11.6557-6567.1990. J Bacteriol. 1990. PMID: 2172216 Free PMC article.
-
The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review.Gene. 1984 Feb;27(2):131-49. doi: 10.1016/0378-1119(84)90135-5. Gene. 1984. PMID: 6327463 Review.
-
Use of transposons to dissect pathogenic strategies of gram-positive bacteria.Methods Enzymol. 1994;235:405-26. doi: 10.1016/0076-6879(94)35158-9. Methods Enzymol. 1994. PMID: 8057913 Review. No abstract available.
Cited by
-
An evolutionarily conserved metabolite inhibits biofilm formation in Escherichia coli K-12.Nat Commun. 2024 Nov 21;15(1):10079. doi: 10.1038/s41467-024-54501-w. Nat Commun. 2024. PMID: 39572562 Free PMC article.
-
The Pseudomonas aeruginosa Resistome: Permanent and Transient Antibiotic Resistance, an Overview.Methods Mol Biol. 2024;2721:85-102. doi: 10.1007/978-1-0716-3473-8_7. Methods Mol Biol. 2024. PMID: 37819517
-
Comprehensive Mapping of EZ-Tn5 Transposon Insertion Sites in Pseudomonas argentinensis SA190 Using RATE-PCR.Bio Protoc. 2025 Jul 20;15(14):e5389. doi: 10.21769/BioProtoc.5389. eCollection 2025 Jul 20. Bio Protoc. 2025. PMID: 40741398 Free PMC article.
-
Improved site-specific mutagenesis in Rhodococcus opacus using a novel conditional suicide plasmid.Appl Microbiol Biotechnol. 2022 Nov;106(21):7129-7138. doi: 10.1007/s00253-022-12204-6. Epub 2022 Oct 4. Appl Microbiol Biotechnol. 2022. PMID: 36194264 Free PMC article.
-
The lipoprotein NlpD in Cronobacter sakazakii responds to acid stress and regulates macrophage resistance and virulence by maintaining membrane integrity.Virulence. 2021 Dec;12(1):415-429. doi: 10.1080/21505594.2020.1870336. Virulence. 2021. PMID: 33459158 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

