Non-canonical Drosophila X chromosome dosage compensation and repressive topologically associated domains
- PMID: 30355339
- PMCID: PMC6199721
- DOI: 10.1186/s13072-018-0232-y
Non-canonical Drosophila X chromosome dosage compensation and repressive topologically associated domains
Abstract
Background: In animals with XY sex chromosomes, X-linked genes from a single X chromosome in males are imbalanced relative to autosomal genes. To minimize the impact of genic imbalance in male Drosophila, there is a dosage compensation complex (MSL) that equilibrates X-linked gene expression with the autosomes. There are other potential contributions to dosage compensation. Hemizygous autosomal genes located in repressive chromatin domains are often derepressed. If this homolog-dependent repression occurs on the X, which has no pairing partner, then derepression could contribute to male dosage compensation.
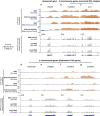
Results: We asked whether different chromatin states or topological associations correlate with X chromosome dosage compensation, especially in regions with little MSL occupancy. Our analyses demonstrated that male X chromosome genes that are located in repressive chromatin states are depleted of MSL occupancy; however, they show dosage compensation. The genes in these repressive regions were also less sensitive to knockdown of MSL components.
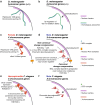
Conclusions: Our results suggest that this non-canonical dosage compensation is due to the same transacting derepression that occurs on autosomes. This mechanism would facilitate immediate compensation during the evolution of sex chromosomes from autosomes. This mechanism is similar to that of C. elegans, where enhanced recruitment of X chromosomes to the nuclear lamina dampens X chromosome expression as part of the dosage compensation response in XX individuals.
Keywords: Dosage compensation; Drosophila melanogaster; Lamina-associated domain; MSL complex; Topologically associated domain.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

