Harnessing longitudinal information to identify genetic variation in tolerance of pigs to Porcine Reproductive and Respiratory Syndrome virus infection
- PMID: 30355341
- PMCID: PMC6201485
- DOI: 10.1186/s12711-018-0420-z
Harnessing longitudinal information to identify genetic variation in tolerance of pigs to Porcine Reproductive and Respiratory Syndrome virus infection
Abstract
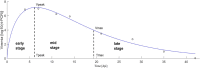
Background: High resistance (the ability of the host to reduce pathogen load) and tolerance (the ability to maintain high performance at a given pathogen load) are two desirable host traits for producing animals that are resilient to infections. For Porcine Reproductive and Respiratory Syndrome (PRRS), one of the most devastating swine diseases worldwide, studies have identified substantial genetic variation in resistance of pigs, but evidence for genetic variation in tolerance has so far been inconclusive. Resistance and tolerance are usually considered as static traits. In this study, we used longitudinal viremia measurements of PRRS virus infected pigs to define discrete stages of infection based on viremia profile characteristics. These were used to investigate host genetic effects on viral load (VL) and growth at different stages of infection, to quantify genetic variation in tolerance at these stages and throughout the entire 42-day observation period, and to assess whether the single nucleotide polymorphism (SNP) WUR10000125 (WUR) with known large effects on resistance confers significant differences in tolerance.
Results: Genetic correlations between resistance and growth changed considerably over time. Individuals that expressed high genetic resistance early in infection tended to grow slower during that time-period, but were more likely to experience lower VL and recovery in growth by the later stage. The WUR genotype was most strongly associated with VL at early- to mid-stages of infection, and with growth at mid- to late-stages of infection. Both, single-stage and repeated measurements random regression models identified significant genetic variation in tolerance. The WUR SNP was significantly associated only with the overall tolerance slope fitted through all stages of infection, with the genetically more resistant AB pigs for the WUR SNP being also more tolerant to PRRS.
Conclusions: The results suggest that genetic selection for improved tolerance of pigs to PRRS is possible in principle, but may be feasible only with genomic selection, requiring intense recording schemes that involve repeated measurements to reliably estimate genetic effects. In the absence of such records, consideration of the WUR genotype in current selection schemes appears to be a promising strategy to improve simultaneously resistance and tolerance of growing pigs to PRRS.
Figures
References
-
- Plastow GS. Genomics to benefit livestock production: improving animal health. Rev Bras Zootec. 2016;45:349–354. doi: 10.1590/S1806-92902016000600010. - DOI
-
- Hermesch S, Dominik S, editors. Breeding focus 2014 – Improving resilience. Armidale: University of New England; 2014.
-
- Morris CA, Bisset SA, Vlassoff A, Wheeler M, West CJ, Devantier BP, et al. Selecting for resilience in Romney sheep under nematode parasite challenge, 1994-2007. New Zeal J Agric Res. 2010;53:245–261. doi: 10.1080/00288233.2010.500714. - DOI
MeSH terms
Grants and funding
- BB/J004235/1 (ISP1)/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20002172 (ISP2.1)/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/30002275 (IPS3.1)/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004235/1 (ISP1)/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20002172 (ISP2.1)/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/30002275 (IPS3.1)/Biotechnology and Biological Sciences Research Council/United Kingdom
- NematodeSystemHealth/Marie Curie Initial Training Networks (FP7-People-2010-ITN)
- KB-12-006.03-004-ASG-LR/Netherlands, and Dutch Ministry of Economic Affairs, Agriculture, and Innovation
- KB-12-006.03-005-ASG-LR/Netherlands, and Dutch Ministry of Economic Affairs, Agriculture, and Innovation
- 2008-55620-19132/National Pork Board
- 2010-65205-20433/USDA NIFA
- 2013-68004-20362/USDA NIFA
- 8042 32000 102/ARS project
LinkOut - more resources
Full Text Sources

