Structures of translationally inactive mammalian ribosomes
- PMID: 30355441
- PMCID: PMC6226290
- DOI: 10.7554/eLife.40486
Structures of translationally inactive mammalian ribosomes
Abstract
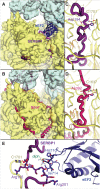
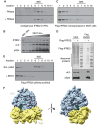
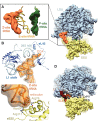
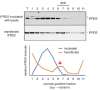
The cellular levels and activities of ribosomes directly regulate gene expression during numerous physiological processes. The mechanisms that globally repress translation are incompletely understood. Here, we use electron cryomicroscopy to analyze inactive ribosomes isolated from mammalian reticulocytes, the penultimate stage of red blood cell differentiation. We identify two types of ribosomes that are translationally repressed by protein interactions. The first comprises ribosomes sequestered with elongation factor 2 (eEF2) by SERPINE mRNA binding protein 1 (SERBP1) occupying the ribosomal mRNA entrance channel. The second type are translationally repressed by a novel ribosome-binding protein, interferon-related developmental regulator 2 (IFRD2), which spans the P and E sites and inserts a C-terminal helix into the mRNA exit channel to preclude translation. IFRD2 binds ribosomes with a tRNA occupying a noncanonical binding site, the 'Z site', on the ribosome. These structures provide functional insights into how ribosomal interactions may suppress translation to regulate gene expression.
Keywords: electron cryomicroscopy; in vitro reconstitution; molecular biophysics; none; ribosome; structural biology.
© 2018, Brown et al.
Conflict of interest statement
AB, MB, MY, JM, SS No competing interests declared
Figures







References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

