Jumonji Inhibitors Overcome Radioresistance in Cancer through Changes in H3K4 Methylation at Double-Strand Breaks
- PMID: 30355483
- PMCID: PMC6245670
- DOI: 10.1016/j.celrep.2018.09.081
Jumonji Inhibitors Overcome Radioresistance in Cancer through Changes in H3K4 Methylation at Double-Strand Breaks
Abstract
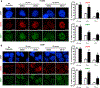
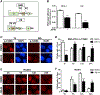
We have uncovered a role for Jumonji inhibitors in overcoming radioresistance through KDM5B inhibition. Pharmacological blockade of Jumonji demethylases with JIB-04 leads to specific accumulation of H3K4me3 at sites marked by γH2AX and impaired recruitment of DNA repair factors, preventing resolution of damage and resulting in robust sensitization to radiation therapy. In DNA-repair-proficient cancer cells, knockdown of the H3K4me3 demethylase KDM5B, but not other Jumonji enzymes, mimics pharmacological inhibition, and KDM5B overexpression rescues this phenotype and increases radioresistance. The H3K4me3 demethylase inhibitor PBIT also sensitizes cancer cells to radiation, while an H3K27me3 demethylase inhibitor does not. In vivo co-administration of radiation with JIB-04 significantly prolongs the survival of mice with tumors even long after cessation of treatment. In human patients, lung squamous cell carcinomas highly expressing KDM5B respond poorly to radiation. Thus, we propose the use of Jumonji KDM inhibitors as potent radiosensitizers.
Keywords: DNA repair; H3K4me3; JARID; JIB-04; Jumonji KDM; KDM5B; lung cancer; radiation therapy; radioresistance; radiosensitization.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures



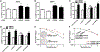

References
-
- Bayo J, Dalvi MP, and Martinez ED (2015). Successful strategies in the discovery of small-molecule epigenetic modulators with anticancer potential. Future Med. Chem 7, 2243–2261. - PubMed
-
- Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, and Kramer MF (2006). High-throughput real-time quantitative reverse transcription PCR. Curr. Protoc. Mol. Biol Chapter 15, Unit 15.18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

