Ranking noncanonical 5' splice site usage by genome-wide RNA-seq analysis and splicing reporter assays
- PMID: 30355602
- PMCID: PMC6280755
- DOI: 10.1101/gr.235861.118
Ranking noncanonical 5' splice site usage by genome-wide RNA-seq analysis and splicing reporter assays
Abstract
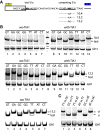
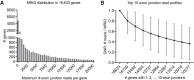
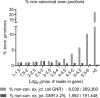
Most human pathogenic mutations in 5' splice sites affect the canonical GT in positions +1 and +2, leading to noncanonical dinucleotides. On the other hand, noncanonical dinucleotides are observed under physiological conditions in ∼1% of all human 5'ss. It is therefore a challenging task to understand the pathogenic mutation mechanisms underlying the conditions under which noncanonical 5'ss are used. In this work, we systematically examined noncanonical 5' splice site selection, both experimentally using splicing competition reporters and by analyzing a large RNA-seq data set of 54 fibroblast samples from 27 subjects containing a total of 2.4 billion gapped reads covering 269,375 exon junctions. From both approaches, we consistently derived a noncanonical 5'ss usage ranking GC > TT > AT > GA > GG > CT. In our competition splicing reporter assay, noncanonical splicing was strictly dependent on the presence of upstream or downstream splicing regulatory elements (SREs), and changes in SREs could be compensated by variation of U1 snRNA complementarity in the competing 5'ss. In particular, we could confirm splicing at different positions (i.e., -1, +1, +5) of a splice site for all noncanonical dinucleotides "weaker" than GC. In our comprehensive RNA-seq data set analysis, noncanonical 5'ss were preferentially detected in weakly used exon junctions of highly expressed genes. Among high-confidence splice sites, they were 10-fold overrepresented in clusters with a neighboring, more frequently used 5'ss. Conversely, these more frequently used neighbors contained only the dinucleotides GT, GC, and TT, in accordance with the above ranking.
© 2018 Erkelenz et al.; Published by Cold Spring Harbor Laboratory Press.
Figures






Similar articles
-
Profiling of cis- and trans-acting factors supporting noncanonical splice site activation.RNA Biol. 2021 Jan;18(1):118-130. doi: 10.1080/15476286.2020.1798111. Epub 2020 Aug 5. RNA Biol. 2021. PMID: 32693676 Free PMC article.
-
Modeling splicing outcome by combining 5'ss strength and splicing regulatory elements.Nucleic Acids Res. 2022 Aug 26;50(15):8834-8851. doi: 10.1093/nar/gkac663. Nucleic Acids Res. 2022. PMID: 35947702 Free PMC article.
-
High-throughput analysis revealed mutations' diverging effects on SMN1 exon 7 splicing.RNA Biol. 2019 Oct;16(10):1364-1376. doi: 10.1080/15476286.2019.1630796. Epub 2019 Jun 19. RNA Biol. 2019. PMID: 31213135 Free PMC article.
-
Pick one, but be quick: 5' splice sites and the problems of too many choices.Genes Dev. 2013 Jan 15;27(2):129-44. doi: 10.1101/gad.209759.112. Genes Dev. 2013. PMID: 23348838 Free PMC article. Review.
-
A novel role of U1 snRNP: Splice site selection from a distance.Biochim Biophys Acta Gene Regul Mech. 2019 Jun;1862(6):634-642. doi: 10.1016/j.bbagrm.2019.04.004. Epub 2019 Apr 28. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 31042550 Free PMC article. Review.
Cited by
-
Alternative splicing modulation by G-quadruplexes.Nat Commun. 2022 May 3;13(1):2404. doi: 10.1038/s41467-022-30071-7. Nat Commun. 2022. PMID: 35504902 Free PMC article.
-
Animal, Fungi, and Plant Genome Sequences Harbor Different Non-Canonical Splice Sites.Cells. 2020 Feb 18;9(2):458. doi: 10.3390/cells9020458. Cells. 2020. PMID: 32085510 Free PMC article.
-
The Experimentally Obtained Functional Impact Assessments of 5' Splice Site GT'GC Variants Differ Markedly from Those Predicted.Curr Genomics. 2020 Jan;21(1):56-66. doi: 10.2174/1389202921666200210141701. Curr Genomics. 2020. PMID: 32655299 Free PMC article.
-
Counteracting the Common Shwachman-Diamond Syndrome-Causing SBDS c.258+2T>C Mutation by RNA Therapeutics and Base/Prime Editing.Int J Mol Sci. 2023 Feb 16;24(4):4024. doi: 10.3390/ijms24044024. Int J Mol Sci. 2023. PMID: 36835434 Free PMC article.
-
Understanding WT1 Alterations and Expression Profiles in Hematological Malignancies.Cancers (Basel). 2023 Jul 4;15(13):3491. doi: 10.3390/cancers15133491. Cancers (Basel). 2023. PMID: 37444601 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
