A combined computational and experimental approach reveals the structure of a C/EBPβ-Spi1 interaction required for IL1B gene transcription
- PMID: 30355733
- PMCID: PMC6311527
- DOI: 10.1074/jbc.RA118.005627
A combined computational and experimental approach reveals the structure of a C/EBPβ-Spi1 interaction required for IL1B gene transcription
Abstract
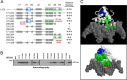
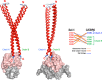
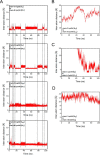
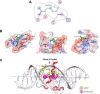
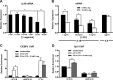
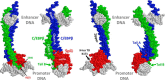
We previously reported that transcription of the human IL1B gene, encoding the proinflammatory cytokine interleukin 1β, depends on long-distance chromatin looping that is stabilized by a mutual interaction between the DNA-binding domains (DBDs) of two transcription factors: Spi1 proto-oncogene at the promoter and CCAAT enhancer-binding protein (C/EBPβ) at a far-upstream enhancer. We have also reported that the C-terminal tail sequence beyond the C/EBPβ leucine zipper is critical for its association with Spi1 via an exposed residue (Arg-232) located within a pocket at one end of the Spi1 DNA-recognition helix. Here, combining in vitro interaction studies with computational docking and molecular dynamics of existing X-ray structures for the Spi1 and C/EBPβ DBDs, along with the C/EBPβ C-terminal tail sequence, we found that the tail sequence is intimately associated with Arg-232 of Spi1. The Arg-232 pocket was computationally screened for small-molecule binding aimed at IL1B transcription inhibition, yielding l-arginine, a known anti-inflammatory amino acid, revealing a potential for disrupting the C/EBPβ-Spi1 interaction. As evaluated by ChIP, cultured lipopolysaccharide (LPS)-activated THP-1 cells incubated with l-arginine had significantly decreased IL1B transcription and reduced C/EBPβ's association with Spi1 on the IL1B promoter. No significant change was observed in direct binding of either Spi1 or C/EBPβ to cognate DNA and in transcription of the C/EBPβ-dependent IL6 gene in the same cells. These results support the notion that disordered sequences extending from a leucine zipper can mediate protein-protein interactions and can serve as druggable targets for regulating gene promoter activity.
Keywords: CCAAT-enhancer–binding protein (C/EBP); ETS transcription factor family; cytokine; gene transcription; inflammation; interleukin 1 (IL-1); molecular docking; molecular dynamics; protein–protein interaction.
© 2018 Pulugulla et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Adamik J., Wang K. Z., Unlu S., Su A. J., Tannahill G. M., Galson D. L., O'Neill L. A., and Auron P. E. (2013) Distinct mechanisms for induction and tolerance regulate the immediate early genes encoding interleukin 1β and tumor necrosis factor α. PLoS One 8, e70622 10.1371/journal.pone.0070622 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

