Clostridioides difficile uses amino acids associated with gut microbial dysbiosis in a subset of patients with diarrhea
- PMID: 30355801
- PMCID: PMC6537101
- DOI: 10.1126/scitranslmed.aam7019
Clostridioides difficile uses amino acids associated with gut microbial dysbiosis in a subset of patients with diarrhea
Abstract
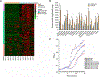
The gut microbiota plays a critical role in pathogen defense. Studies using antibiotic-treated mice reveal mechanisms that increase susceptibility to Clostridioides difficile infection (CDI), but risk factors associated with CDI in humans extend beyond antibiotic use. Here, we studied the dysbiotic gut microbiota of a subset of patients with diarrhea and modeled the gut microbiota of these patients by fecal transplantation into germ-free mice. When challenged with C. difficile, the germ-free mice transplanted with fecal samples from patients with dysbiotic microbial communities showed increased gut amino acid concentrations and greater susceptibility to CDI. A C. difficile mutant that was unable to use proline as an energy source was unable to robustly infect germ-free mice transplanted with a dysbiotic or healthy human gut microbiota. Prophylactic dietary intervention using a low-proline or low-protein diet in germ-free mice colonized by a dysbiotic human gut microbiota resulted in decreased expansion of wild-type C. difficile after challenge, suggesting that amino acid availability might be important for CDI. Furthermore, a prophylactic fecal microbiota transplant in mice with dysbiosis reduced proline availability and protected the mice from CDI. Last, we identified clinical risk factors that could potentially predict gut microbial dysbiosis and thus greater susceptibility to CDI in a retrospective cohort of patients with diarrhea. Identifying at-risk individuals and reducing their susceptibility to CDI through gut microbiota-targeted therapies could be a new approach to preventing C. difficile infection in susceptible patients.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures
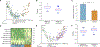
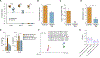


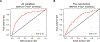
References
-
- Schulz MD, Atay C, Heringer J, Romrig FK, Schwitalla S, Aydin B, Ziegler PK, Varga J, Reindl W, Pommerenke C, Salinas-Riester G, Böck A, Alpert C, Blaut M, Polson SC, Brandl L, Kirchner T, Greten FR, Polson SW, Arkan MC, High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature 514, 508–512 (2014). - PMC - PubMed
-
- Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, Hirota K, Matsushita M, Furuta Y, Narazaki M, Sakaguchi N, Kayama H, Nakamura S, Iida T, Saeki Y, Kumanogoh A, Sakaguchi S, Takeda K, Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 68, 2646–2661 (2016). - PubMed
-
- Rajilić-Stojanović M, Jonkers DM, Salonen A, Hanevik K, Raes J, Jalanka J, de Vos WM, Manichanh C, Golic N, Enck P, Philippou E, Iraqi FA, Clarke G, Spiller RC, Penders J, Intestinal microbiota and diet in IBS: Causes, consequences, or epiphenomena? Am. J. Gastroenterol. 110, 278–287 (2015). - PMC - PubMed
-
- Keeney KM, Yurist-Doutsch S, Arrieta MC, Finlay BB, Effects of antibiotics on human microbiota and subsequent disease. Annu. Rev. Microbiol 68, 217–235 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

