Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus
- PMID: 30355968
- PMCID: PMC6267276
- DOI: 10.3390/v10110581
Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus
Abstract
Extreme resistance (ER) is a type of R-gene-mediated resistance that rapidly induces a symptomless resistance phenotype, which is different from the phenotypical R-resistance manifested by the programmed cell death, accumulation of reactive oxygen species, and hypersensitive response. The Rsv3 gene in soybean cultivar L29 is responsible for ER against the avirulent strain G5H of soybean mosaic virus (SMV), but is ineffective against the virulent strain G7H. Rsv3-mediated ER is achieved through the rapid accumulation of callose, which arrests SMV-G5H at the point of infection. Callose accumulation, however, may not be the lone mechanism of this ER. Analyses of RNA-seq data obtained from infected soybean plants revealed a rapid induction of the abscisic acid pathway at 8 h post infection (hpi) in response to G5H but not to G7H, which resulted in the down-regulation of transcripts encoding β-1,3 glucanases that degrade callose in G5H-infected but not G7H-infected plants. In addition, parts of the autophagy and the small interfering (si) RNA pathways were temporally up-regulated at 24 hpi in response to G5H but not in response to G7H. The jasmonic acid (JA) pathway and many WRKY factors were clearly up-regulated only in G7H-infected plants. These results suggest that ER against SMV-G5H is achieved through the quick and temporary induction of ABA, autophagy, and the siRNA pathways, which rapidly eliminate G5H. The results also suggest that suppression of the JA pathway in the case of G5H is important for the Rsv3-mediated ER.
Keywords: Rsv3; extreme resistance; signal transduction pathways; soybean mosaic virus.
Conflict of interest statement
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures
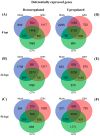
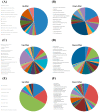


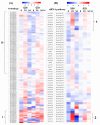

References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

