Genetic divergence of HIV-1 B subtype in Italy over the years 2003-2016 and impact on CTL escape prevalence
- PMID: 30356083
- PMCID: PMC6200748
- DOI: 10.1038/s41598-018-34058-7
Genetic divergence of HIV-1 B subtype in Italy over the years 2003-2016 and impact on CTL escape prevalence
Abstract
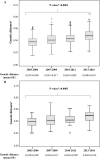
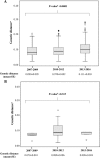
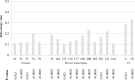
HIV-1 is characterized by high genetic variability, with implications for spread, and immune-escape selection. Here, the genetic modification of HIV-1 B subtype over time was evaluated on 3,328 pol and 1,152 V3 sequences belonging to B subtype and collected from individuals diagnosed in Italy between 2003 and 2016. Sequences were analyzed for genetic-distance from consensus-B (Tajima-Nei), non-synonymous and synonymous rates (dN and dS), CTL escapes, and intra-host evolution over four time-spans (2003-2006, 2007-2009, 2010-2012, 2013-2016). Genetic-distance increased over time for both pol and V3 sequences (P < 0.0001 and 0.0003). Similar results were obtained for dN and dS. Entropy-value significantly increased at 16 pol and two V3 amino acid positions. Seven of them were CTL escape positions (protease: 71; reverse-transcriptase: 35, 162, 177, 202, 207, 211). Sequences with ≥3 CTL escapes increased from 36.1% in 2003-2006 to 54.0% in 2013-2016 (P < 0.0001), and showed better intra-host adaptation than those containing ≤2 CTL escapes (intra-host evolution: 3.0 × 10-3 [2.9 × 10-3-3.1 × 10-3] vs. 4.3 × 10-3 [4.0 × 10-3-5.0 × 10-3], P[LRT] < 0.0001[21.09]). These data provide evidence of still ongoing modifications, involving CTL escape mutations, in circulating HIV-1 B subtype in Italy. These modifications might affect the process of HIV-1 adaptation to the host, as suggested by the slow intra-host evolution characterizing viruses with a high number of CTL escapes.
Conflict of interest statement
The authors have no financial and non-financial competing interests that might be perceived to influence the results and/or discussion reported in this paper. However, Francesca Ceccherini-Silberstein reports personal fees from Gilead Sciences, Bristol-Myers Squibb, Abbvie, Roche Diagnostics, Janssen-Cilag, Abbott Molecular, ViiV Healthcare; grants and personal fees from Merck Sharp & Dohme; grants from Italian Ministry of Education, University and Research (MIUR). Carlo Federico Perno reports grants from Italian Ministry of Instruction, University and Research (MIUR), and from Aviralia Foundation; personal fees from Gilead Sciences, Abbvie, Roche Diagnostics, Janssen-Cilag, Abbott Molecular; and grants and personal fees from Bristol-Myers Squibb, Merck Sharp & Dohme, and ViiV Healthcare. All other authors have nothing to declare.
Figures