T-bet controls intestinal mucosa immune responses via repression of type 2 innate lymphoid cell function
- PMID: 30356098
- PMCID: PMC6548562
- DOI: 10.1038/s41385-018-0092-6
T-bet controls intestinal mucosa immune responses via repression of type 2 innate lymphoid cell function
Abstract
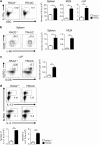
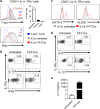
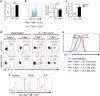
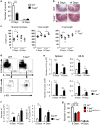
Innate lymphoid cells (ILCs) play an important role in regulating immune responses at mucosal surfaces. The transcription factor T-bet is crucial for the function of ILC1s and NCR+ ILC3s and constitutive deletion of T-bet prevents the development of these subsets. Lack of T-bet in the absence of an adaptive immune system causes microbiota-dependent colitis to occur due to aberrant ILC3 responses. Thus, T-bet expression in the innate immune system has been considered to dampen pathogenic immune responses. Here, we show that T-bet plays an unexpected role in negatively regulating innate type 2 responses, in the context of an otherwise intact immune system. Selective loss of T-bet in ILCs leads to the expansion and increased activity of ILC2s, which has a functionally important impact on mucosal immunity, including enhanced protection from Trichinella spiralis infection and inflammatory colitis. Mechanistically, we show that T-bet controls the intestinal ILC pool through regulation of IL-7 receptor signalling. These data demonstrate that T-bet expression in ILCs acts as the key transcriptional checkpoint in regulating pathogenic vs. protective mucosal immune responses, which has significant implications for the understanding of the pathogenesis of inflammatory bowel diseases and intestinal infections.
Conflict of interest statement
The authors declare no competing interests.
Figures