Mechanoresponsive stem cells acquire neural crest fate in jaw regeneration
- PMID: 30356216
- PMCID: PMC6481292
- DOI: 10.1038/s41586-018-0650-9
Mechanoresponsive stem cells acquire neural crest fate in jaw regeneration
Abstract
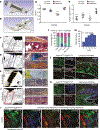
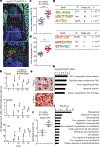
During both embryonic development and adult tissue regeneration, changes in chromatin structure driven by master transcription factors lead to stimulus-responsive transcriptional programs. A thorough understanding of how stem cells in the skeleton interpret mechanical stimuli and enact regeneration would shed light on how forces are transduced to the nucleus in regenerative processes. Here we develop a genetically dissectible mouse model of mandibular distraction osteogenesis-which is a process that is used in humans to correct an undersized lower jaw that involves surgically separating the jaw bone, which elicits new bone growth in the gap. We use this model to show that regions of newly formed bone are clonally derived from stem cells that reside in the skeleton. Using chromatin and transcriptional profiling, we show that these stem-cell populations gain activity within the focal adhesion kinase (FAK) signalling pathway, and that inhibiting FAK abolishes new bone formation. Mechanotransduction via FAK in skeletal stem cells during distraction activates a gene-regulatory program and retrotransposons that are normally active in primitive neural crest cells, from which skeletal stem cells arise during development. This reversion to a developmental state underlies the robust tissue growth that facilitates stem-cell-based regeneration of adult skeletal tissue.
Conflict of interest statement
Figures













References
-
- Ilizarov GA The tension-stress effect on the genesis and growth of tissues: part II. The influence of the rate and frequency of distraction. Clin. Orthop. Relat. Res 239, 263–285 (1989). - PubMed
-
- Ilizarov GA The tension-stress effect on the genesis and growth of tissues. Part I. The influence of stability of fixation and soft-tissue preservation. Clin. Orthop. Relat. Res 238, 249–281 (1989). - PubMed
-
- Tahiri Y, Viezel-Mathieu A, Aldekhayel S, Lee J & Gilardino M The effectiveness of mandibular distraction in improving airway obstruction in the pediatric population. Plast. Reconstr. Surg 133, 352e–359e (2014). - PubMed
-
- McCarthy JG, Schreiber J, Karp N, Thorne CH & Grayson BH Lengthening the human mandible by gradual distraction. Plast. Reconstr. Surg 89, 1–8 (1992). - PubMed
-
- Khansa I et al. Airway and feeding outcomes of mandibular distraction, tongue-lip adhesion, and conservative management in Pierre Robin sequence: a prospective study. Plast. Reconstr. Surg 139, 975e–983e (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

