Recombination and intraspecific polymorphism for the presence and absence of entire chromosomes in mitochondrial genomes
- PMID: 30356223
- PMCID: PMC6461862
- DOI: 10.1038/s41437-018-0153-3
Recombination and intraspecific polymorphism for the presence and absence of entire chromosomes in mitochondrial genomes
Abstract
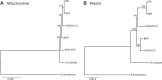
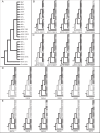
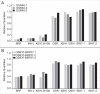
Although mitochondrial genomes are typically thought of as single circular molecules, these genomes are fragmented into multiple chromosomes in many eukaryotes, raising intriguing questions about inheritance and (in)stability of mtDNA in such systems. A previous comparison of mitochondrial genomes from two different individuals of the angiosperm species Silene noctiflora found variation in the presence of entire mitochondrial chromosomes. Here, we expand on this work with a geographically diverse sampling of 25 S. noctiflora populations and the closely related species S. turkestanica and S. undulata. Using a combination of deep sequencing and PCR-based screening for the presence of 22 different mitochondrial chromosomes, we found extensive variation in the complement of chromosomes across individuals. Much of this variation could be attributed to recent chromosome loss events, suggesting that the massively expanded and fragmented mitochondrial genomes of S. noctiflora may have entered a phase of genome reduction in which they are losing entire chromosomes at a rapid rate. Sequence analysis of mitochondrial and plastid genomes revealed genealogical differences both between these organelles and within the mitochondrial genome, indicating a history of recombination. Evidence that recombination has generated novel combinations of alleles was more frequent between loci on different mitochondrial chromosomes than it was within chromosomes. Therefore, the fragmentation of mitochondrial genomes and the assortment of chromosomes during mitochondrial inheritance appears to have contributed to a history of sexual-like recombination in the mtDNA of this species.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
The massive mitochondrial genome of the angiosperm Silene noctiflora is evolving by gain or loss of entire chromosomes.Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10185-91. doi: 10.1073/pnas.1421397112. Epub 2015 May 5. Proc Natl Acad Sci U S A. 2015. PMID: 25944937 Free PMC article.
-
Recent acceleration of plastid sequence and structural evolution coincides with extreme mitochondrial divergence in the angiosperm genus Silene.Genome Biol Evol. 2012;4(3):294-306. doi: 10.1093/gbe/evs006. Epub 2012 Jan 12. Genome Biol Evol. 2012. PMID: 22247429 Free PMC article.
-
Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates.PLoS Biol. 2012 Jan;10(1):e1001241. doi: 10.1371/journal.pbio.1001241. Epub 2012 Jan 17. PLoS Biol. 2012. PMID: 22272183 Free PMC article.
-
[Prokaryotic and Mitochondrial Linear Genomes: Their Genesis, Evolutionary Significance, and the Problem of Replicating Chromosome Ends].Mol Biol (Mosk). 2019 Mar-Apr;53(2):218-224. doi: 10.1134/S0026898419020125. Mol Biol (Mosk). 2019. PMID: 31099772 Review. Russian.
-
One ring to rule them all? Genome sequencing provides new insights into the 'master circle' model of plant mitochondrial DNA structure.New Phytol. 2013 Dec;200(4):978-85. doi: 10.1111/nph.12395. New Phytol. 2013. PMID: 24712049 Review.
Cited by
-
Organelle Genomes of Epipogium roseum Provide Insight into the Evolution of Mycoheterotrophic Orchids.Int J Mol Sci. 2024 Jan 27;25(3):1578. doi: 10.3390/ijms25031578. Int J Mol Sci. 2024. PMID: 38338856 Free PMC article.
-
Highly active repeat-mediated recombination in the mitogenome of the aquatic grass Hygroryza aristata.BMC Plant Biol. 2024 Jul 8;24(1):644. doi: 10.1186/s12870-024-05331-x. BMC Plant Biol. 2024. PMID: 38973002 Free PMC article.
-
De novo Assembly and Comparative Analyses of Mitochondrial Genomes in Piperales.Genome Biol Evol. 2023 Mar 3;15(3):evad041. doi: 10.1093/gbe/evad041. Genome Biol Evol. 2023. PMID: 36896589 Free PMC article.
-
A complete mitochondrial genome for fragrant Chinese rosewood (Dalbergia odorifera, Fabaceae) with comparative analyses of genome structure and intergenomic sequence transfers.BMC Genomics. 2021 Sep 18;22(1):672. doi: 10.1186/s12864-021-07967-7. BMC Genomics. 2021. PMID: 34536995 Free PMC article.
-
Assembly and comparative analysis of the first complete mitochondrial genome of Astragalus membranaceus (Fisch.) Bunge: an invaluable traditional Chinese medicine.BMC Plant Biol. 2024 Nov 8;24(1):1055. doi: 10.1186/s12870-024-05780-4. BMC Plant Biol. 2024. PMID: 39511474 Free PMC article.
References
-
- Arrieta-Montiel MP, Mackenzie SA (2011). Plant Mitochondrial Genomes and Recombination BT - Plant Mitochondria. In: Kempken F (ed) Springer New York: New York, NY, pp 65–82.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

