Milk microbiome diversity and bacterial group prevalence in a comparison between healthy Holstein Friesian and Rendena cows
- PMID: 30356246
- PMCID: PMC6200206
- DOI: 10.1371/journal.pone.0205054
Milk microbiome diversity and bacterial group prevalence in a comparison between healthy Holstein Friesian and Rendena cows
Abstract
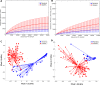
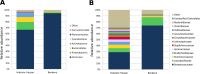
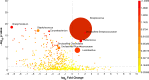
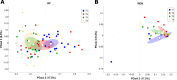
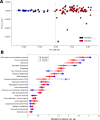
Dry and early lactation periods represent the most critical phases for udder health in cattle, especially in highly productive breeds, such as the Holstein Friesian (HF). On the other hand, some autochthonous cattle breeds, such as the Rendena (REN), have a lower prevalence of mastitis and other transition-related diseases. In this study, milk microbiota of 6 HF and 3 REN cows, all raised on the same farm under the same conditions, was compared. A special focus was placed on the transition period to define bacterial groups' prevalence with a plausible effect on mammary gland health. Four time points (dry-off, 1 d, 7-10 d and 30 d after calving) were considered. Through 16S rRNA sequencing, we characterized the microbiota composition for 117 out of the 144 milk samples initially collected, keeping only the healthy quarters, in order to focus on physiological microbiome changes and avoid shifts due to suspected diseases. Microbial populations were very different in the two breeds along all the time points, with REN milk showing a significantly lower microbial biodiversity. The taxonomic profiles of both cosmopolitan and local breeds were dominated by Firmicutes, mostly represented by the Streptococcus genus, although in very different proportions (HF 27.5%, REN 68.6%). Large differences in HF and REN cows were, also, evident from the metabolic predictive analysis from microbiome data. Finally, only HF milk displayed significant changes in the microbial composition along the transition period, while REN maintained a more stable microbiota. In conclusion, in addition to the influence on the final characteristics of dairy products obtained from milk of the two breeds, differences in the milk microbiome might, also, have an impact on their mammary gland health.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
What we have lost: Mastitis resistance in Holstein Friesians and in a local cattle breed.Res Vet Sci. 2018 Feb;116:88-98. doi: 10.1016/j.rvsc.2017.11.020. Epub 2017 Nov 29. Res Vet Sci. 2018. PMID: 29223308
-
Changes in bovine milk bacterial microbiome from healthy and subclinical mastitis affected animals of the Girolando, Gyr, Guzera, and Holstein breeds.Int Microbiol. 2022 Nov;25(4):803-815. doi: 10.1007/s10123-022-00267-4. Epub 2022 Jul 15. Int Microbiol. 2022. PMID: 35838927
-
Composition of the teat canal and intramammary microbiota of dairy cows subjected to antimicrobial dry cow therapy and internal teat sealant.J Dairy Sci. 2018 Nov;101(11):10191-10205. doi: 10.3168/jds.2018-14858. Epub 2018 Aug 29. J Dairy Sci. 2018. PMID: 30172408
-
Review of the bacterial composition of healthy milk, mastitis milk and colostrum in small ruminants.Res Vet Sci. 2021 Nov;140:1-5. doi: 10.1016/j.rvsc.2021.07.022. Epub 2021 Jul 27. Res Vet Sci. 2021. PMID: 34358776 Review.
-
The Role of Innate Immune Response and Microbiome in Resilience of Dairy Cattle to Disease: The Mastitis Model.Animals (Basel). 2020 Aug 11;10(8):1397. doi: 10.3390/ani10081397. Animals (Basel). 2020. PMID: 32796642 Free PMC article. Review.
Cited by
-
Amplicon-sequencing of raw milk microbiota: impact of DNA extraction and library-PCR.Appl Microbiol Biotechnol. 2021 Jun;105(11):4761-4773. doi: 10.1007/s00253-021-11353-4. Epub 2021 Jun 1. Appl Microbiol Biotechnol. 2021. PMID: 34059942 Free PMC article.
-
Characterization of Bacterial Microbiota Composition along the Gastrointestinal Tract in Rabbits.Animals (Basel). 2020 Dec 26;11(1):31. doi: 10.3390/ani11010031. Animals (Basel). 2020. PMID: 33375259 Free PMC article.
-
Temperature abuse and Salmonella Typhimurium colonization disrupt the indigenous bacterial communities of pasteurized bovine milk over time.Sci Rep. 2025 Jul 2;15(1):22567. doi: 10.1038/s41598-025-06838-5. Sci Rep. 2025. PMID: 40594414 Free PMC article.
-
Elucidation of the Bovine Intramammary Bacteriome and Resistome from healthy cows of Swiss dairy farms in the Canton Tessin.Front Microbiol. 2023 Jul 31;14:1183018. doi: 10.3389/fmicb.2023.1183018. eCollection 2023. Front Microbiol. 2023. PMID: 37583512 Free PMC article.
-
Milk Microbiota: What Are We Exactly Talking About?Front Microbiol. 2020 Feb 14;11:60. doi: 10.3389/fmicb.2020.00060. eCollection 2020. Front Microbiol. 2020. PMID: 32117107 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

